Figures & data
Figure 1. In vitro release of GEM from RGD-BSANP-GEM in PBS containing 10% (v/v) fetal bovine serum (FBS) at 37°C.
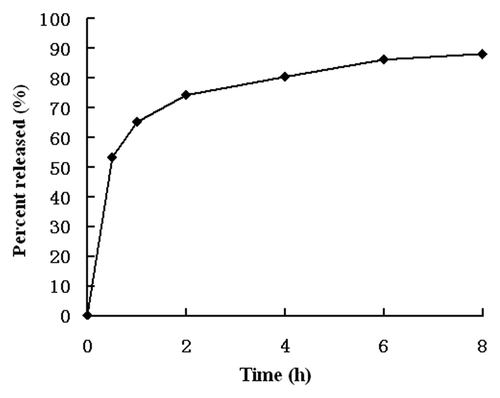
Figure 2. (A) After dissolved in PBS of pH 8.5 and kept in 4°C for 4 weeks, the sample dose not aggregate and still appears milky white colloidal solution. (B) Four weeks later.
Figure 3. Flow cytometric analysis of surface-expressed integrin αvβ3 receptor in BxPC-3, SW1990, PANC-1 and CFPAC-1 cells. Histograms depict relative fluorescence intensity (log scale) of control (above) and integrin αvβ3 (below). BxPC-3 cells showed the highest expression of αvβ3 compared with the other three cells [The positive rate of integrin αvβ3 receptor on the four cell lines: (A) 86.94%, (B) 71.50%, (C) 13.58%, (D) 12.51%].
![Figure 3. Flow cytometric analysis of surface-expressed integrin αvβ3 receptor in BxPC-3, SW1990, PANC-1 and CFPAC-1 cells. Histograms depict relative fluorescence intensity (log scale) of control (above) and integrin αvβ3 (below). BxPC-3 cells showed the highest expression of αvβ3 compared with the other three cells [The positive rate of integrin αvβ3 receptor on the four cell lines: (A) 86.94%, (B) 71.50%, (C) 13.58%, (D) 12.51%].](/cms/asset/1d3a35f4-afba-448d-8436-de43087e14ec/kcbt_a_10918692_f0003.gif)
Figure 4. Fluorescein isothiocyanate (FITC) was labeled onto the nanoparticles to visualize the uptake of nanoparticles. Fluorescent signal was observed directly under the fluorescence microscope with an unaided eye. The signal of RGD-conjugated BSANPs treated cells (B) was higher than that of BSANPs without RGD conjugation (A).
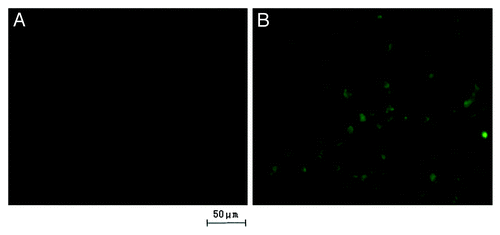
Figure 5. Concentration dependence of nanoparticles uptake by BxPC-3 cells. With the increase of incubation concentration of nanoparticles, the intensity of fluorescence enhanced gradually. The nanoparticles were incubated with BxPC-3 cells for 16 h. The intensity of fluorescence of RGD-conjugated BSANPs was significantly higher than that of BSANPs without RGD conjugation at different concentration points (n = 3, *p < 0.05).
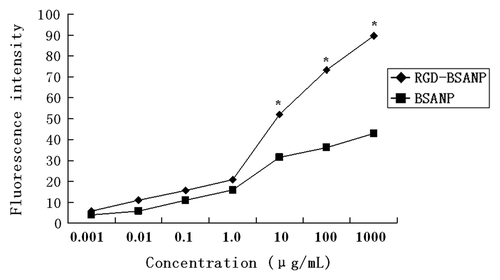
Figure 6. Time-dependence of nanoparticles uptake by BxPC-3 cells. The concentration of nanoparticles was 10 μg/mL. The intensity of fluorescence increased as time went on. Cells treated with FITC labeled RGD-conjugated BSANPs showed higher fluorescence intensity than cells treated with BSANPs without RGD conjugation (*p < 0.05).
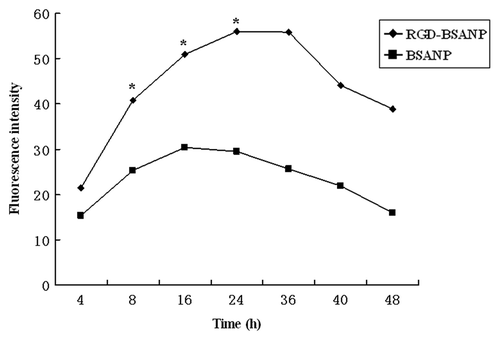
Figure 7. Inhibition of RGD-conjugated BSANPs uptake by free RGD peptides. Excessive free RGD peptides were added into the cell samples 3 h prior to the incubation with RGD-conjugated BSANPs for another 6 h at room temperature. Cellular uptake of RGD-conjugated BSANPs was inhibited by free RGD peptide.
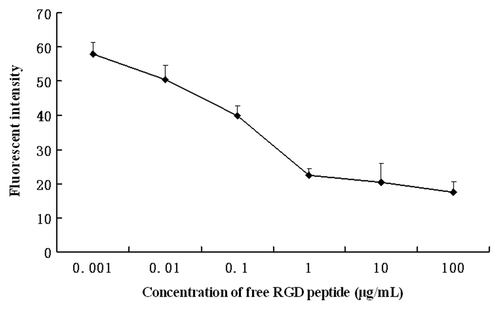
Figure 8. Confocal image of BxPC-3 cells incubated with FITC labeled RGD-conjugated BSANPs. Nanoparticles in BxPC-3 cells were shown by green fluorescence. Nuclei were shown in blue. Nanoparticles The nanoparticles were found to be located close to the nuclei.
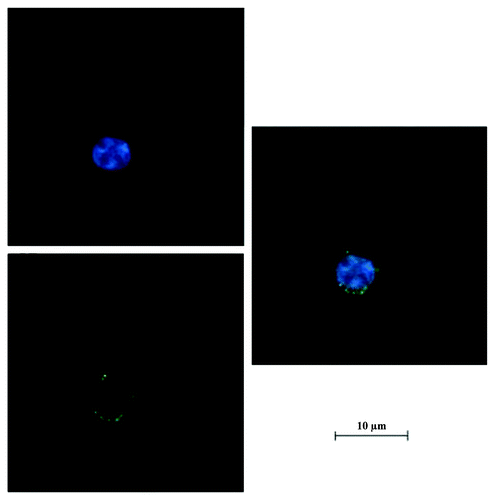
Figure 9. Cytotoxicity of RGD-conjugated BSANPs by MTT assay. Cell viability was not significantly decreased as the exposure time and concentration increased. No significant difference was found between the RGD-conjugated BSANPs and BSANPs (p > 0.05).
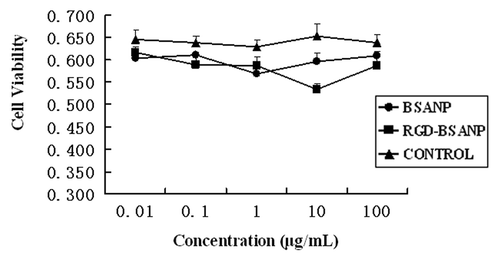
Figure 10. Analysis of cell viability treated with different agents. Compared with other groups, the in vitro antitumor efficacy was improved by targeting gemcitabine-loaded nanoparticles to BxPC-3 cells using RGD peptides,*p < 0.05, vs. BSANP-GEM and GEM group. (A) 48 h, (B) 72 h.
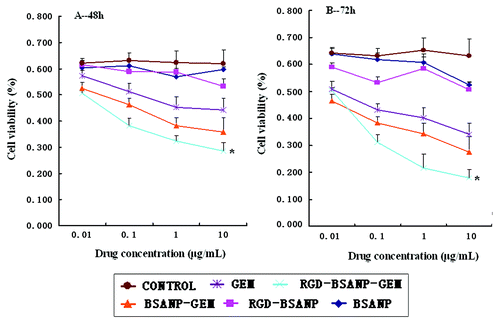
Figure 11. Cell viability of the four pancreatic cancer cells treated with RGD-conjugated gemcitabine-loaded nanoparticles. Among the four pancreatic cancer cells, BxPC-3 cells acquired the lowest cell viability (*p < 0.05).
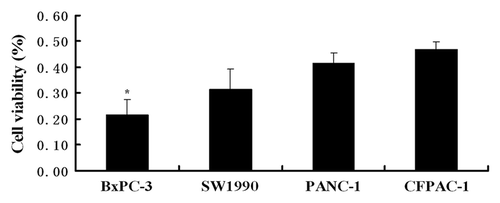
Table 1. Cell cycle distribution and apoptotic index of BxPC-3 with different treatments
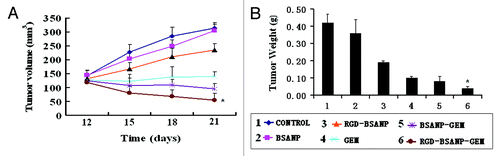
Figure 12. Changes in the tumor volume and weight after treatment in nude mice bearing xenografts of human pancreatic cancer with saline (control, 1), BSANP alone (2), RGD-BSANP conjugate (3), GEM alone (90 mg/kg, 4), BSANP-GEM (90 mg/kg GEM equivalent, 5) and RGD-BSANP-GEM (90 mg/kg GEM equivalent, 6). The in vivo tumor growth was significantly reduced by treatment with RGD-BSANP-GEM (*p < 0.05).