Figures & data
Figure 1. eIF4E expression is elevated in human NSCLC cell lines (A) and tissues (B and C). (A) Whole-cell protein lysates were extracted from the indicated normal and NSCLC cell lines and used for detection of eIF4E expression with western blotting. (B and C) eIF4E expression in human NSCLC tissues was detected with IHC and scored as positive or negative expression (B). The representative images were also presented (C). SCC, squamous cell carcinoma; AD, adenocarcinoma; Normal, adjacent normal lung tissue.
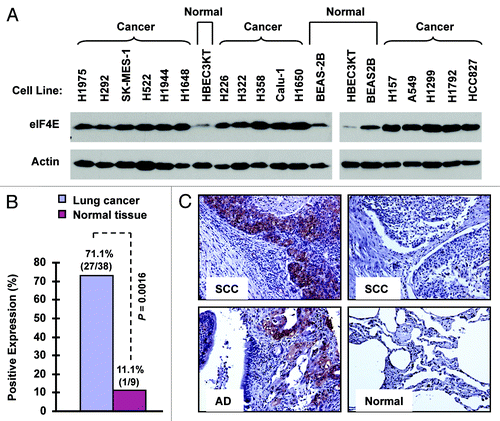
Figure 2. Knockdown of eIF4E (A, D and E) inhibits the growth of NSCLC cells (B and C) and induces apoptosis (D) with suppression of cap-dependent translation (E). (A and B) The indicated NSCLC cell lines were transfected with control (Ctrl) or eIF4E siRNA (20 nM) for 48 h and then subjected to western blot analysis for detection of eIF4E (A). The cells were also re-plated in 96-well plates. Cell numbers were estimated every 24 h with the SRB assay (B). The data are means ± SDs of four replicates. (C) 801-C and 801-D cells were transfected with control or eIF4E siRNA for overnight and equal numbers of cells were then used for soft agar in 35 mm diameter Petri dishes. After 14 d, colony numbers were counted and averaged from five random microscopic field or view. The final data are means ± SDs of triplicate independent determinations. The Student t-test was used to compare growth-inhibitory effects between two groups. (D and E) The indicated cell lines were transfected with control or eIF4E siRNA for 72 h (D) or 48 h (E). The cells were then harvested for preparation of whole-cell protein lysates and subsequent western blotting for detection of the given proteins. In (D) the control siRNA-transfected cells were exposed to 50 ng/ml tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) for 20 h before harvesting the cells. UD, undetected.
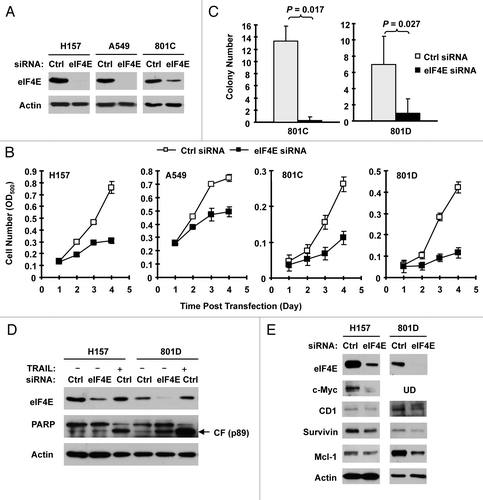
Figure 3. eIF4E expression is increased in metastatic NSCLC cells (A) and tissues (B) and is associated with cell invasion (C and D). (A) eIF4E expression in 801C and 801D cells was detected with western blot analysis. (B) eIF4E expression in primary and matched metastatic NSCLC tissues was detected with IHC. (C and D) Both 801C and 801D cells were transfected with control (Ctrl) or eIF4E siRNA for 48 h and then subjected to matrigel chamber invasion assay. After 36 h, the non-invaded cells and collagen matrix on top of the membranes were removed. Invasive cells on the bottoms of the membranes were counted and normalized by the live cells cultured under the same conditions (C). Representative images of invasive cells on the membranes were also shown (D). The data are means ± SDs of triplicate determinations. The student t-test was used to compare inhibitory effects on invasion between two groups.
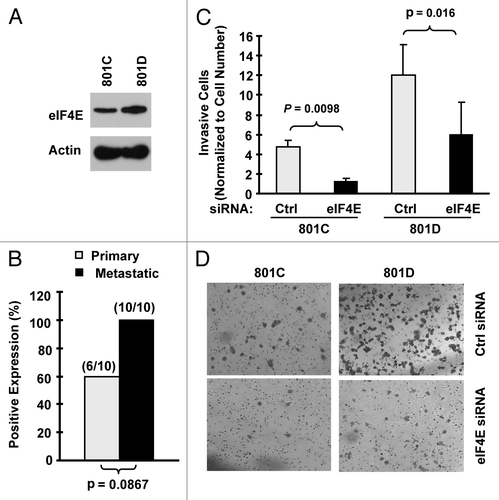
Figure 4. Erlotinib-resistant NSCLC cells possess elevated levels of eIF4E protein (A and B) and mRNA (C and D). (A and B) Whole-cell protein lysates were prepared from the indicated cell lines (A) or cell lines exposed to different concentrations of erlotinib for 6 h (B) and then used for western blot analysis to detect the given proteins as indicated. (C) Total cellular RNA was isolated from both parental and HCC827/ER cells for detection of eIF4E mRNA by RT-PCR. (D) eIF4E promoter activities in the given cell lines were performed with transfection of the given reporter constructs into HCC827 or HCC827/ER cells followed with a luciferase activity assay after 48 h. Each column represents the mean ± SD of triplicate determinations.
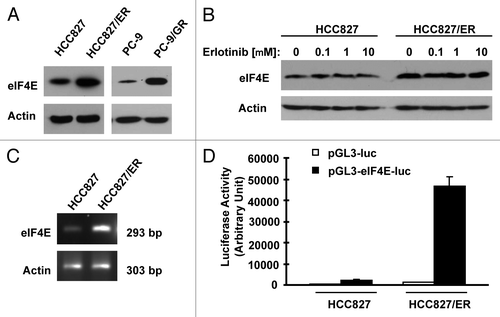
Figure 5. Erlotinib-resistance cells exhibit elevated eIF4F assembly (A) and expression of oncogenic proteins regulated by the cap-dependent translation (B). (A) Whole-cell protein lysates prepared from the given cell lines were used for m7GTP pull-down assay followed with western blot analysis to detect the indicated proteins. LE, long exposure. (B) Whole-cell protein lysates were prepared from the indicated cell lines and then used for western blot analysis to detect the given proteins as indicated.
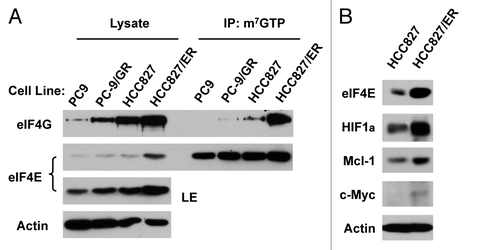
Figure 6. Inhibition of eIF4F formation by knocking down eIF4E (A) or eIF4G (C) or by inhibiting eIF4E and eIF4G interaction with 4EGI-1 (B) sensitizes erlotinib-resistant cells to erlotinib. (A) HCC828/ER cells were transfected with control (Ctrl) or eIF4E siRNA for overnight and then exposed to 2 μM erlotinib for 3 d. B, HCC828/ER cells were treated with indicated concentrations of erlotinib in the absence and presence of 4EGI-1 for 3 d. (C) HCC828/ER cells were transfected with control (Ctrl) or eIF4G siRNA for overnight and then exposed to the indicated concentrations of erlotinib for 3 d. After the aforementioned treatments, the cell numbers were estimated by the SRB assay. The data are means ± SDs of four replicate determinations. The numbers by the lines in (B) are combination indexes for the combinations of erlotinib and 4EGI-1.
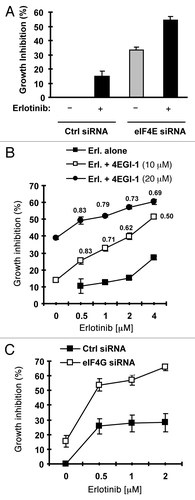
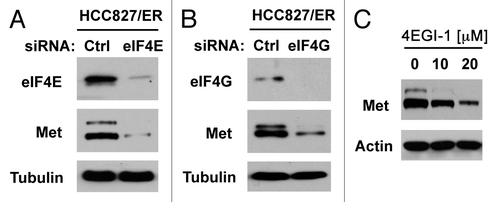
Figure 7. Inhibition of eIF4F formation by knocking down eIF4E (A) or eIF4G (B) or by inhibiting eIF4E and eIF4G interaction with 4EGI-1 (C) reduces c-Met protein levels. (A and B) HCC828/ER cells were transfected with control (Ctrl), eIF4E or eIF4G siRNA for 48 h. (C) HCC828/ER cells were treated with the indicated concentrations of 4EGI-1 for 24 h. After the aforementioned treatments, the cells were harvested for preparation of whole-cell protein lysates and subsequent western blotting.