Figures & data
Table 2. List of deregulated miRNAs at > 1.5-fold in A549/CDDP vs. A549 cells
Figure 1. Comparison between microarray and qRT-PCR. The bars indicate the fold change which was determined for the expression ratio of A549/CDDP vs. A549. Results from all seven comparisons are consistent.
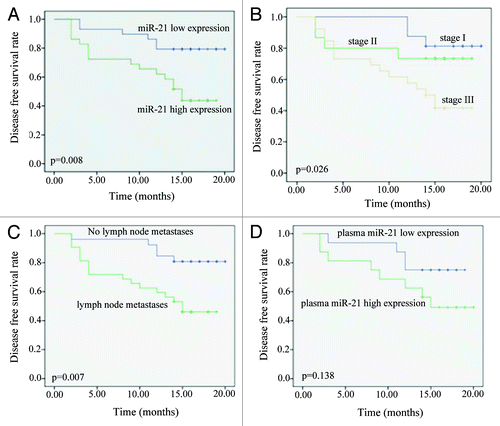
Figure 2. Effects of miR-21 on drug sensitivity of NSCLC cells. (A) Effects of anti-miR-21 on A549/CDDP cells. Transfection of the A549/CDDP cells with anti-miR-21 increases their sensitivity to platinum treatment. (B) Effects of pre-miR-21 on A549 cells. Transfection of the A549 cells with pre-miR-21 decreases their sensitivity to platinum treatment.
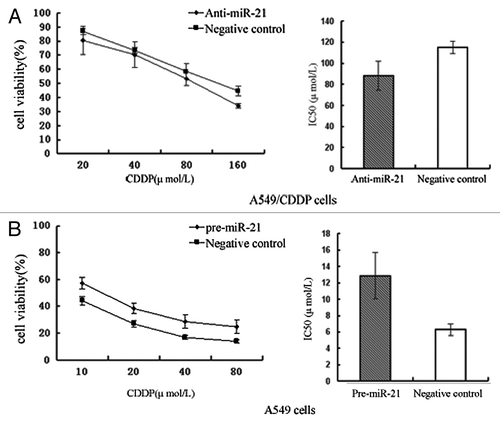
Table 3. Comparison of several clinicopathologic factors and miR-21 expression level in 58 NSCLC patients undergone adjuvant platinum based chemotherapy
Figure 3. Kaplan-Meier estimation of Disease Free Survival time (DFS) of NSCLC. (A) DFS analysis of patients according to the miR-21 relative expression. The expression levels of miR-21 in 58 patients were measured by Real-time PCR. High expression is based on the median values of miR-21 relative expression. High miR-21 expression (p = 0.008) had a significant relationship with patient’s shorter DFS. (B) DFS analysis of patients according to their clinical stage. Patients were classified as either early or late clinical stage. Clinical stage had a significant relationship with patient’s DFS (p = 0.026). (C) DFS analysis of patients according to their lymph node status. Patients classified as either lymph node metastasis positive or negative. Lymph node metastasis was found to be strongly associated with poor outcome (p = 0.007). Log-rank p values are from Kaplan-Meier analysis. (D) DFS analysis of patients according to the miR-21 relative expression in plasma samples of 32 patients. The increased expression level of miR-21 in plasma is marginally associated with shorter DFS (p = 0.138).
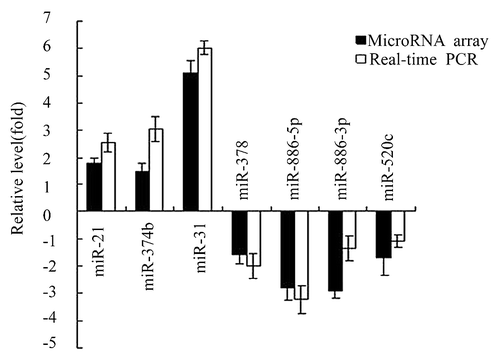
Table 4. Univariate and multivariate analysis for factors related to DFS using the COX proportional hazard model
Table 5. Correlation of miR-21 expression levels between plasma samples and corresponding primary tumor tissue samples
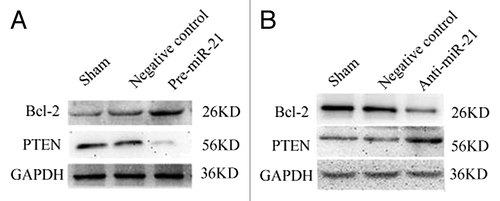
Figure 4. Expression of PTEN and BCL-2 in the A549 and A549/CDDP cells assessed by western blot after transfected with either scrambled or pre-miR-21 and anti-miR -21 expression. (A) The effects of miR-21 on the expression of PTEN and BCL-2 in A549 cells. Transfection of A549 cells with pre-miR-21 resulted in a decrease of PTEN levels and an increase of Bcl-2. (B) The effects of miR-21 on the expression of PTEN and BCL-2 in A549/CDDP cells. Transfection of A549/CDDP cells with anti-miR-21 resulted in an increase of PTEN levels and a decrease of Bcl-2. Sham: A549 or A549/CDDP without transfection; Negative control: A549 or A549/CDDP with transfection of Anti-miR and Pre-miR.