Figures & data
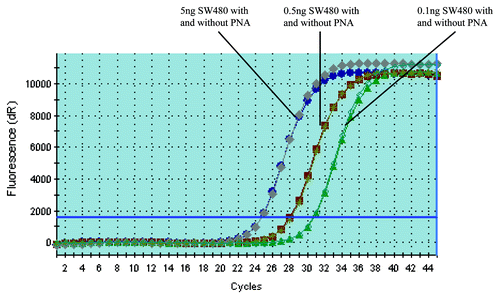
Figure 1. PNA doesn’t affect the amplification of KRAS mutant DNA. PCR with and without PNA have almost the same amplification curves when the template is KRAS mutant DNA (The PCR amplification curve with and without PNA are almost totally overlapping each other). The amount of SW480 DNA added is 5, 0.5 and 0.1 ng, respectively (corresponding to the amplification curve from left to right in the figure).
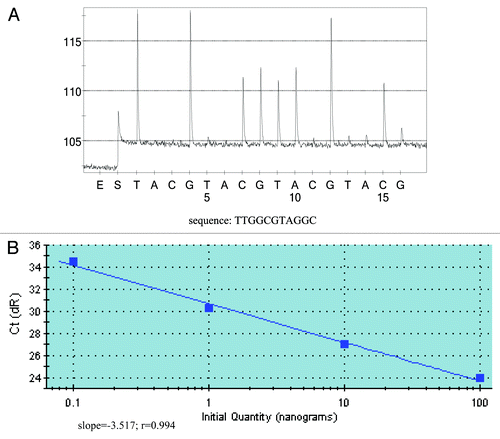
Figure 2. Detection sensivitity of PNA-PCR assay and quantification of KRAS mutant DNA. (A) Detection sensitivity of PNA-PCR assay. The sequencing primer used was PF1, and as little as 20 pg of mutant DNA can be detected from 100 ng KRAS wild-type DNA (1:5,000). The sequencing pyrogram shows the absence of wild-type sequence, which indicates that PNA could successfully suppress the amplification of wild-type sequences. (B) Quantification of KRAS mutant DNA. Varying amounts of KRAS mutant DNA were plotted against Ct values (threshold cycle). Slope, r value and regression line are shown.
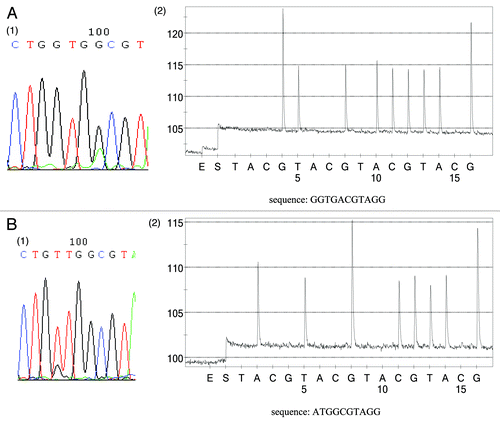
Table 1. KRAS mutation status of FFPE tissues and plasma samples
Table 2. Comparison of KRAS mutation rate between plasma and tumor tissue
Figure 3. Comparison of KRAS status between FFPE tumor tissues and corresponding plasma samples. (A) Subject (15) has the same KRAS alternation (G13D) between tumor tissue and plasma sample. A1 KRAS status of FFPE tumor tissue. A2 KRAS status of plasma sample using sequencing primer PF2. (B) Inconsistency between plasma sample and corresponding tumor tissue (subject Y3). Tumor tissue contains a G12V mutation (1) whereas plasma sample carries a G12D mutation (sequencing primer PF1) (2). It should be noted that the grams of pyrosequencing only reveal the KRAS mutant sequence, while no wild-type sequence was observed, which also proved that only KRAS mutant alleles could be amplified in this assay.