Figures & data
Figure 1. Expression of G-CSF and G-CSFR in human glioma samples. (A) RT-PCR results revealed G-CSF and G-CSFR mRNA expression in human glioma samples; mRNA expression level of G-CSF and G-CSFR was undetected in normal brain tissue (NB). (B) Relative mRNA levels in grade I/II gliomas (n = 7), grade III gliomas (n = 8), grade IV gliomas (n = 12), and normal brain tissue (NB, n = 5). (C) Western blot assay shows expression of G-CSFR in glioma samples. Expression of G-CSFR was undetected in normal brain tissue (NB). (D) Prevalence of G-CSF and G-CSFR expression in grade I/II gliomas (7/8), grade III gliomas (8/9), grade IV gliomas (12/12), and normal brain tissue (NB, 0/5).
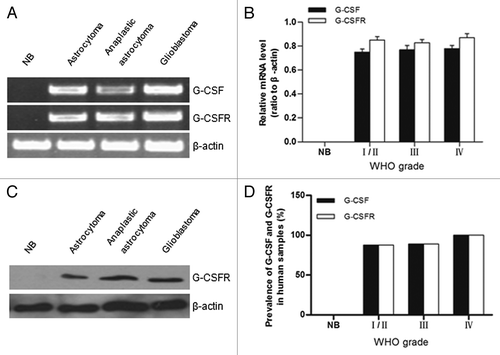
Figure 2. Immunohistochemical analysis of G-CSFR expression in human glioma samples and normal brain tissue. (A) Immunohistochemistry shows G-CSFR expression is mainly localized in the cytoplasm (b, e, and h, brown; arrow) of glioma cells. G-CSFR is widely expressed in glioma cells (a, d, and g), but not in the normal brain tissue (NB, j). c, f, i, and l are negative controls for grade I/II glioma, grade III glioma, grade IV glioma, and normal brain tissue (NB), respectively. For negative controls, cells were similarly processed as the tested groups but without the primary antibody. (B) Average G-CSFR-positive cells in grade I/II gliomas (80.71% ± 5.06%), grade III gliomas (79.50% ± 6.07%), grade IV gliomas (82.36% ± 6.68%), and normal brain tissue (0%). The difference between grades was not statistically significant (p > 0.05).
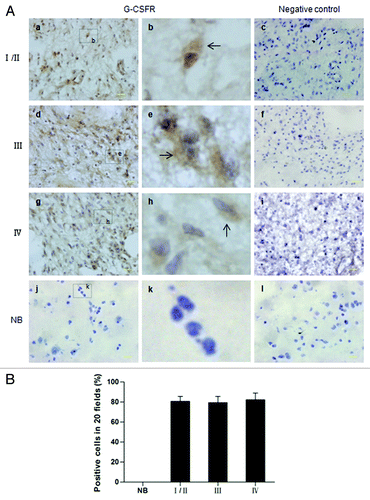
Figure 3. Expression of G-CSF and G-CSFR in glioma cell cultures. (A) Shows G-CSFR immunofluorescence (red) in glioma cell culture derived from glioma sample (G0518) and glioma cell lines (T98G, U251, and U87), but not in primary cultured astrocytes. Cell nuclei were counterstained with DAPI (blue). Magnification: 200 × scale: 100 μm. (B) Western blot shows G-CSFR protein expression in glioma cells and primary astrocytes. (C) RT-PCR shows mRNA expression levels of G-CSF and G-CSFR in glioma cells: G0518, and T98G cells, but not in primary astrocytes.
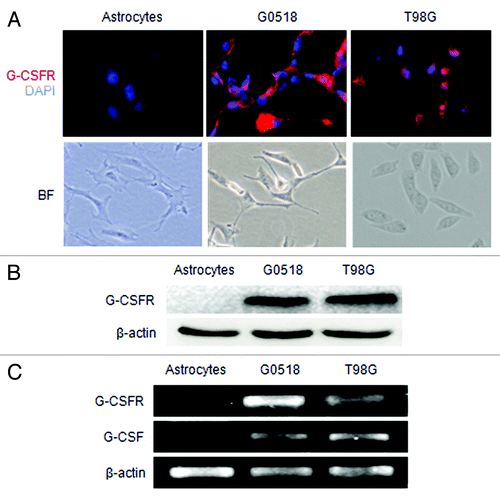
Figure 4. Effect of G-CSF on cell viability and proliferation of glioma cells. (A and B) Cell viability was determined by MTT assay. (A) Showing dose dependent effect of G-CSF on glioma cells; (B) Showing time dependent effect of G-CSF on glioma cells. Data are expressed as ratio to the control value (without G-CSF). (C, D) BrdU assay shows that the percentage of BrdU positive cells is significantly increased in G-CSF treated groups compared with groups without G-CSF treatment. (E, F) Colony formation assay shows that the G-CSF treated cells formed more colonies compared with those without G-CSF treatment. (*p < 0.05, #p < 0.01; each experimental point was performed with three wells, and values represent mean ± SD of three independent experiments).
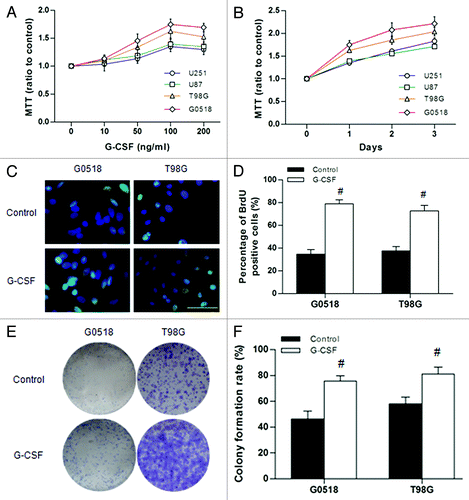
Figure 5. Effects of G-CSFR antibody in glioma cells. (A, B) MTT and BrdU incorporation assays show G-CSFR antibody decreases cell viability and growth, respectively. (C, D, and E) Flow cytometry assay revealed G-CSFR antibody significantly increased Annexin-V-positive cells in glioma cells, indicating that G-CSFR antibody induced apoptosis. (*p < 0.05, #p < 0.01; each experimental point was performed with three wells, and values represent mean ± SD of three independent experiments).
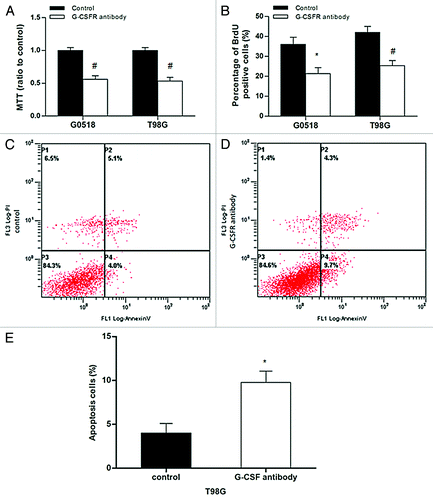
Figure 6. Effects of G-CSF on migration and invasion of glioma cells. (A, B) G-CSF treatment significantly increased migration ability of G0518 and T98G, as demonstrated by increases in migration distance (A) and migration cells (B). G-CSFR antibody inhibited migration ability of glioma cell. (C) Invasion ability of G-CSF treated group is higher than that of untreated group, while that of G-CSFR antibody treated group is decreased. (*p < 0.05, #p < 0.01. Each experimental point was performed with three wells, and values represent mean ± SD of three independent experiments).
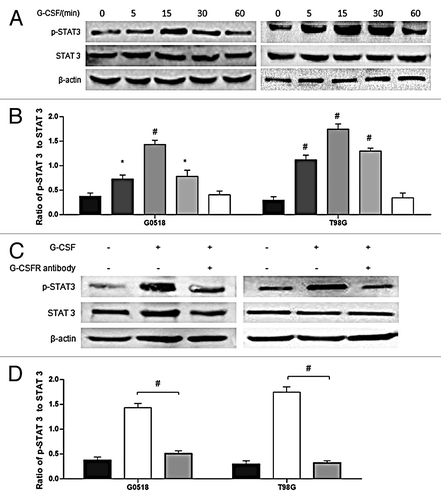
Figure 7. Phosphorylation of STAT 3 is regulated by G-CSF. (A, B) western blot analysis shows time-dependent effects of G-CSF on activation of STAT 3 in G0518 and T98G cells. (C, D) western blot analysis shows that G-CSFR antibody inhibits G-CSF induced elevation of p-STAT 3. (*p < 0.05, #p < 0.01, Each experimental point was performed with three wells, and values represent mean ± SD of three independent experiments).
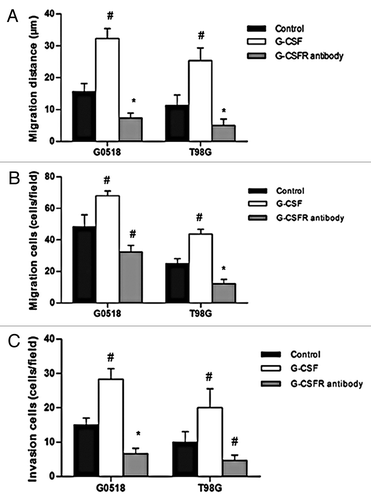
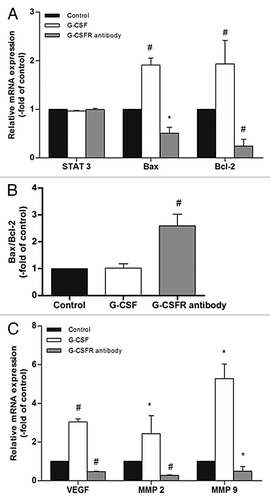
Figure 8. STAT 3 downstream genes are modulated by G-CSF. (A, C) Real time RT-PCR shows neither G-CSF nor G-CSFR antibody affects STAT 3 expression. On the other hand, G-CSF upregulates expression of STAT 3 downstream genes including Bax, Bcl-2, VEGF, MMP 2, and MMP 9. Neutralization of G-CSFR with its antibody inhibits expression of these genes. (B) Ratio of Bax to Bcl-2 is not affected by G-CSF, but is significantly elevated by G-CSFR antibody. (*p < 0.05, #p < 0.01). Each experimental point was performed with three wells, and values represent mean ± SD of three independent experiments).