Figures & data
Figure 1. Luciferase activity of the Gli-reporter gene pT81 co-transfected with expression plasmids for Gli1 and Gli3. Cells from the line T98G were transfected with the pT81 Gli reporter gene together with expression plasmids for Gli1 (hGli1) or Gli3 (hGli3), respectively. Luciferase activity from the Gli reporter plasmid was compared with the activity of the corresponding control plasmid in the presence of the same expression plasmid set to 100%. Error bars indicate standard deviations. For the determination of mean and standard deviation six transfection experiments were performed. **p < 0.01.
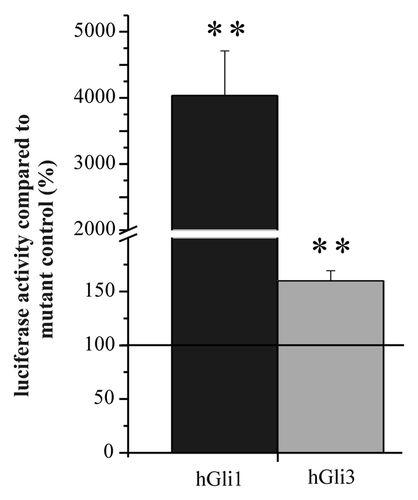
Figure 2. Luciferase expression from the Gli reporter gene after transfection into cells from GBM incubated without and in the presence of 5 and 10 µM cyclopamine. Luciferase activity of cells transfected with the reporter gene “pT81_Gli” was compared with the activity of cells transfected with the corresponding control plasmid “pT81_Gli-Mut” set to 100%. Below the bars the cultures are indicated. Error bars indicate standard deviations. The level of significance for enhanced transcription compared with the control plasmid as determined by student’s t-test is indicated by asterisks. *p < 0.05; **p < 0.005; ***p < 0.0005. For the determination of mean and standard deviations six transfection experiments were performed.
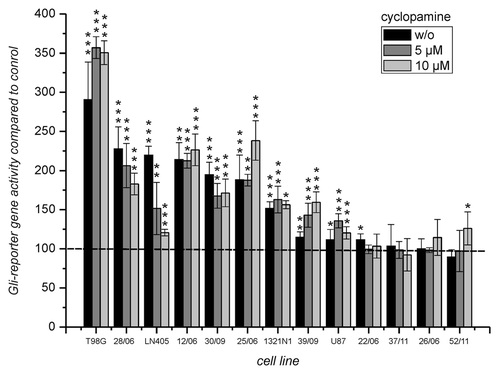
Figure 3. Effect of cyclopamine on the viability of cells derived from GBM. The cells from the experiment in were incubated for 24 h in the presence of different concentrations of cyclopamine (0, 0.1, 0.5, 1, 2.5, 5, 7.5, 10, 15 and 20 µM). Viability was determined by the CellTiterGlo Assay and data was fitted using the Boltzman equation. For comparison viability of cells from untreated control was set to 100%.
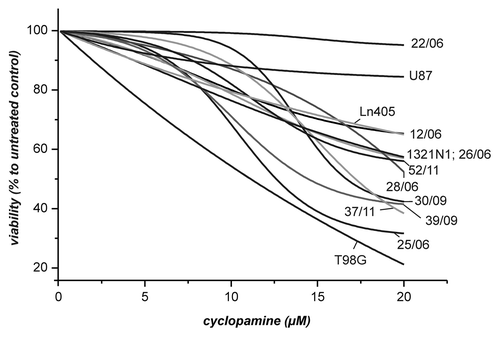
Figure 4. Correlation between reporter gene activity and viability in the presence of 5 and 10 µM cyclopamine. Viability of cells as determined in was compared with the reporter gene activity from (untreated cells). The comparison only includes cells which exhibited at least a reporter gene enhancement above 1.5 x.
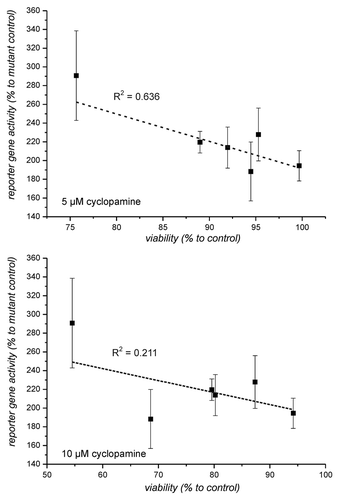
Table 1. Viability of cells in the presence of 1 mM cyclopamine as determined by extrapolation of the fitting curves in
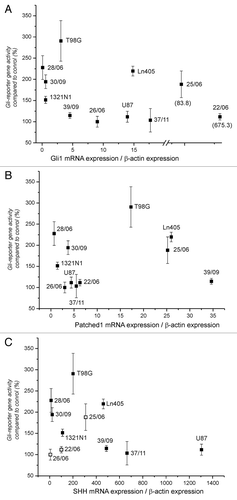
Figure 5. Correlation between expression of Gli1, SHH and Patched1 mRNA and reporter gene enhancement. The relative amount of mRNA was determined by normalization of each mRNA amount of Gli1 (A), Patched1 (B) and SHH (C) and to the corresponding amount of β-actin in each sample. Open squares in part C indicate that the amount of SHH mRNA in these samples were below the used standard curve (less than 100 copies).