Figures & data
Figure 1. Effect of Ginsenoside Rg3 on the cell viability and apoptosis of EPCs. (A) The chemical structure of Ginsenoside Rg3. (B) Endothelial phenotype of outgrowth ECs, derived from isolated cord blood MNCs was confirmed by FACS analysis, demonstrating the expression of endothelial markers CD31, CD34, KDR, CXCR4, c-Kit, CD133. The graph represents the percentage of outgrowth ECs. (C, D) Cytotoxic effect was determined using EPCs and LLCs in the presence of Ginsenoside Rg3. After cultivation for 24 h under normal culture conditions, the total cell number was analyzed using a WST-1 assay. The cell number was not altered significantly by a treatment with up to 600 ng/mL Ginsenoside Rg3, demonstrating that Ginsenoside Rg3 does not affect the viability of EPCs and LLC. (E, F) Apoptosis effect was determined using EPCs and LLCs in the presence of Ginsenoside Rg3. After cultivation for 24 h under serum free culture conditions, cleaved caspase-3, a typical indicator of apoptosis, was increased after treatment with Ginsenoside Rg3 in a dose dependent manner, as demonstrated by western blot analysis.
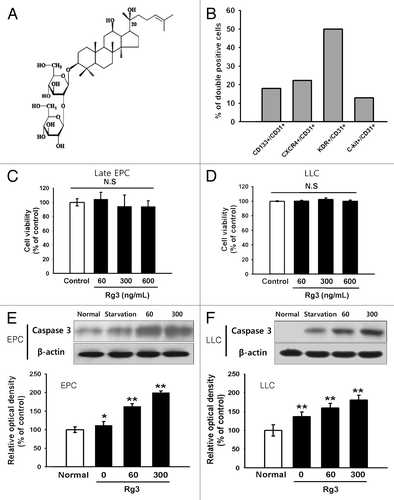
Figure 2. Effect of Ginsenoside Rg3 on the VEGFR2 expression in EPCs. (A) mRNA level of VEGFR2 in Rg3-treated outgrowth ECs, demonstrated by RT-PCR analysis. (B) VEGFR2 protein expression in Rg3-treated outgrowth ECs, demonstrated by western blotting analysis.
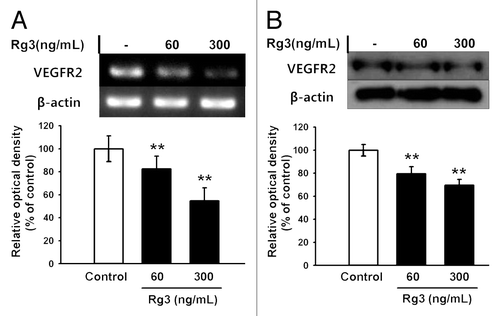
Figure 3. Effect of Ginsenoside Rg3 on the proliferation of EPCs. (A) Effect of Ginsenoside Rg3 on VEGF-induced cell proliferation in outgrowth ECs. Cell proliferation assay was performed at 24 h after treatment with or without Ginsenoside Rg3 in the presence of VEGF. **p < 0.005 (significance between control and VEGF), *p < 0.05 (significance between VEGF and VEGF plus Ginsenoside Rg3). (B) Ginsenoside Rg3 attenuates the phosphorylation of ERK and p38 protein levels but not JNK, as demonstrated by western blotting analysis using the outgrowth ECs. The total ERK, JNK, p38 and β-actin were probed as a protein loading control. All data was obtained from at least three independent experiments. (C) The graph represents the relative optical density of each group via the ERK, p38, JNK control group. The bar graph indicating each percentage was expressed using the untreated groups as 100%. *p < 0.05, **p < 0.005.
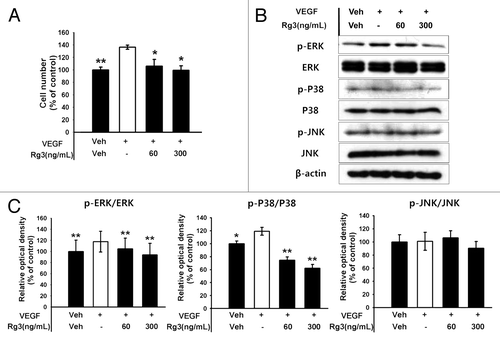
Figure 4. Effect of Ginsenoside Rg3 on cell cycle progression in EPCs. (A) The cell cycle progression was analyzed by western blotting analysis using multiple antibodies against pivotal cell cycle molecules. (B) The graph represents the relative optical density of each group via the β-actin control group. The bar graph indicating each percentage was expressed using the untreated groups as 100%. *p < 0.05, **p < 0.005.
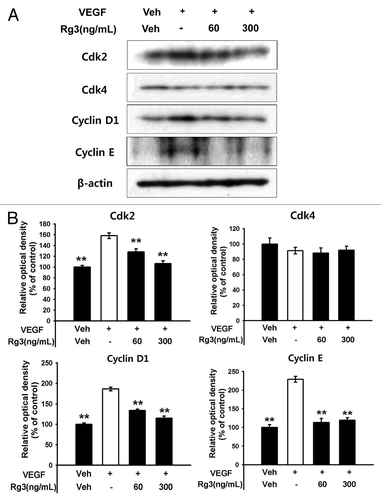
Figure 5. Effect of Ginsenoside Rg3 on the ability of migration of EPCs. (A) Representative optical micrographs showing wound healing by monolayers. Inhibitive effect of Rg3 on VEGF induced outgrowth ECs migration. Ex vivo cultured outgrowth ECs were subjected to wound healing migration assay. Bar, 500 μm. (B) Bar graph represents the number of migrated cells. Fields were chosen randomly from various section levels to ensure the objectivity of sampling. *significance between the control and VEGF, p < 0.05, significance between VEGF and VEGF plus Ginsenoside Rg3, p < 0.05.
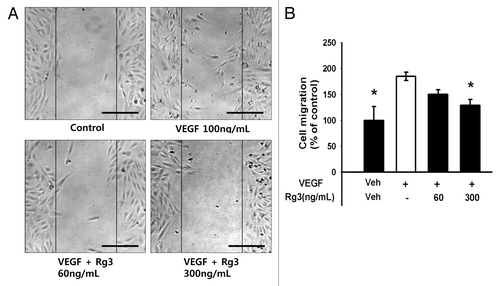
Figure 6. Effect of Ginsenoside Rg3 on the ability of tube formation of EPCs. (A) Representative tubular network structure in the outgrowth ECs with or without Ginsenoside Rg3. Bar, 500 μm. (B, C) Significant differences between Rg3-treated outgrowth ECs and vehicle-treated outgrowth ECs. The bar graph represents the number of intact loops in the capillary networks, as measured by image-J software. The graph represents the length of the tube in the capillary networks (*p < 0.05, **p < 0.005).
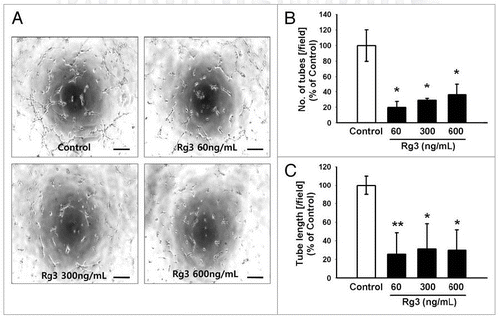
Figure 7. Effect of Ginsenoside Rg3 on tumor angiogenesis and tumor growth. (A) After s.c. injection with 5 × 104 LLC tumor cells, the mice were administered orally with the DDW vehicle (control) or 600 μg/kg Ginsenoside Rg3 daily for 23 d after the initiation of therapy (n = 5). The mice were sacrificed at day 24 because the mice treated with the vehicle only had large tumors. (B) The vehicle or Ginsenoside Rg3 (600 μg/kg) was administered orally everyday using protocols of oral administration injection. Tumor growth was measured with calipers every 3 or 4 d using the formula V = height ×length × depth (mm3). Tumor depth is defined as the horizontal extent of tumor’s lateral part. Bar, 1 cm. There is significant difference between tumor group and Rg3 treated tumor group at day 24. (C) Representative photomicrographs of CD31 capillaries (5~20 μm in diameter) and arterials (150~500 μm in diameter) in tumor sections of LLC bearing mice. Bar, 500 μm. (D, E) The number of CD31-stained capillary expressing arterial and capillary were counted using image J program. The fields were chosen randomly from various section levels to ensure the objectivity of sampling. *p < 0.05.
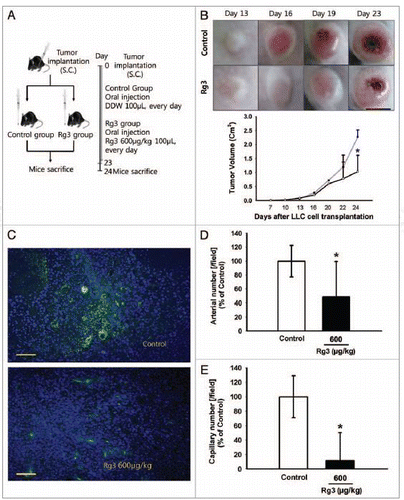
Figure 8. Effect of Ginsenoside Rg3 on EPC mobilization. (A) After s.c. injection with 5 × 104 LLC, the mice were administered orally with DDW vehicle (control) or 600 μg/kg Ginsenoside Rg3 daily for 4 d after the initiation of therapy. The circulating MNCs was harvested at day 5 by Ficoll-gradient centrifugation. (B) Subsequent gating procedure was used to deplete the CD45 hematopoietic cells. Double positive fraction for detecting CD34+VEGFR2+ cells in the gated CD45- cell population (subsets represented in the upper-right quadrant) was expressed as circulating progenitor cells (EPCs) in murine PB. This figure means negative control and positive control. (C) Statistical difference between Ginsenoside after the oral administration of both Rg3 and vehicle in LLC tumor bearing mice (n = 5). The bar graph represents a marked difference in the EPC frequency at day 5 after daily injection of Rg3 or the vehicle. *p < 0.05.
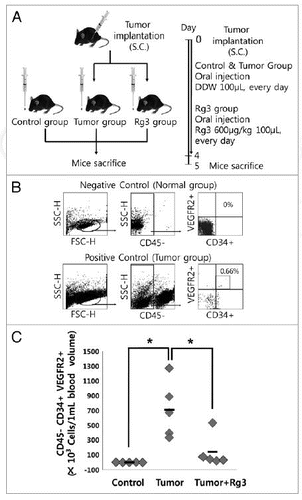
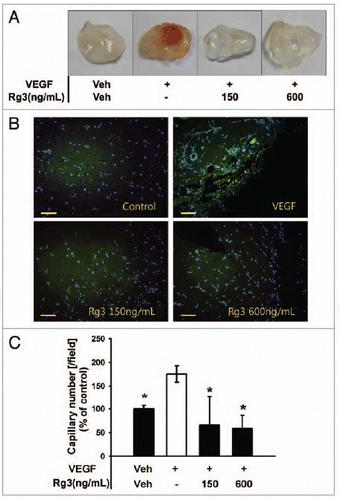
Figure 9.Ginsenoside Rg3 inhibits VEGF-induced angiogenesis in vivo. (A) Experimental protocols of VEGF dependent matrigel plug assay to examine the effect of Ginsenoside Rg3 on neovessel formation. (B) Representative photomicrographs of CD31 stained matrigel sections of mice treated with the vehicle, VEGF (100 ng/mL in matrigel) and various doses of Ginsenoside Rg3 (150 ng/mL and 600 ng/mL) with VEGF (100 ng/mL). Bar, 500 μm. (C) The number of capillaries was counted using the image J program. The fields were chosen randomly from various section levels to ensure the objectivity of sampling. The graph represents the number of CD31 positive capillaries (*p < 0.05). Percent inhibition was expressed using untreated groups as 100% (n = 5).