Figures & data
Figure 1. Cell proliferation is significantly decreased by silencing Rac1 expression in NSCLC cell lines. (A) To assess the silencing of Rac1 expression in NSCLC cell lines, NCI-H1703 and A549 cells were either subjected to mock transfection without siRNA (lane 1) or transfected with nontargeting (NT) siRNA (lane 2) or the indicated Rac1 siRNAs (lanes 3 and 4). Cell lysates were prepared and subjected to ECL-Western blotting using antibodies to GAPDH and Rac1. Results are representative of three independent experiments. (B) Quantitative densitometry was conducted to determine the protein levels of Rac1 in the ECL-Western blots described in A. The values are normalized to densitometric values obtained from ECL-Western blots of cells subjected to NT siRNA transfection. Results are the mean ± SE from three independent experiments. (C) Cell proliferation was assayed by measuring [3H]thymidine uptake 72 h after transfecting the cells with 25 nM of the indicated siRNAs. The values are normalized to [3H]thymidine uptake by cells subjected to NT siRNA transfection. Results are the mean ± SE from three independent experiments conducted with six replicates for each treatment in each experiment. (D) Cell proliferation rate was assayed by measuring [3H]thymidine uptake at the indicated time points 72 h after transfecting the cells with 25 nM of the indicated siRNAs. Results are the mean ± SE from three independent experiments conducted with four replicates for each treatment, in each experiment. Symbols above a column indicate a statistical comparison between the indicated sample and the control sample of cells transfected with NT siRNA (*, p < 0.01)
![Figure 1. Cell proliferation is significantly decreased by silencing Rac1 expression in NSCLC cell lines. (A) To assess the silencing of Rac1 expression in NSCLC cell lines, NCI-H1703 and A549 cells were either subjected to mock transfection without siRNA (lane 1) or transfected with nontargeting (NT) siRNA (lane 2) or the indicated Rac1 siRNAs (lanes 3 and 4). Cell lysates were prepared and subjected to ECL-Western blotting using antibodies to GAPDH and Rac1. Results are representative of three independent experiments. (B) Quantitative densitometry was conducted to determine the protein levels of Rac1 in the ECL-Western blots described in A. The values are normalized to densitometric values obtained from ECL-Western blots of cells subjected to NT siRNA transfection. Results are the mean ± SE from three independent experiments. (C) Cell proliferation was assayed by measuring [3H]thymidine uptake 72 h after transfecting the cells with 25 nM of the indicated siRNAs. The values are normalized to [3H]thymidine uptake by cells subjected to NT siRNA transfection. Results are the mean ± SE from three independent experiments conducted with six replicates for each treatment in each experiment. (D) Cell proliferation rate was assayed by measuring [3H]thymidine uptake at the indicated time points 72 h after transfecting the cells with 25 nM of the indicated siRNAs. Results are the mean ± SE from three independent experiments conducted with four replicates for each treatment, in each experiment. Symbols above a column indicate a statistical comparison between the indicated sample and the control sample of cells transfected with NT siRNA (*, p < 0.01)](/cms/asset/26e15e73-0aaa-4625-8b5f-0933099ddcd5/kcbt_a_10920082_f0001.gif)
Figure 2. Silencing of Rac1 expression in NSCLC cells diminishes colony formation in soft agar. NCI-H1703 cells (A–C and G) and A549 cells (D–F and G) were transfected with the indicated siRNAs and an equal number of live cells were plated 24 h later in soft agar. Digital images of the cells were collected 5 weeks later (A–F), and the numbers of visible colonies in the agar were counted (G). The results in G are the mean ± SE from three independent experiments conducted with triplicate samples. Symbols above a column indicate a statistical comparison between the indicated sample and the control sample of cells transfected with NT siRNA (*p < 0.05).
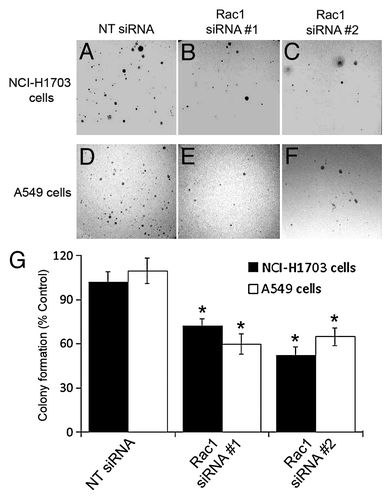
Figure 3. Silencing of Rac1 in NSCLC cells slows progression through the G1 phase of the cell cycle. Changes in cell cycle progression were determined by transfecting NCI-H1703 cells (A) or A549 (B) cells without siRNA (mock transfection) or with the indicated siRNAs and staining the cells with propidium iodide 72 h post-transfection, followed by flow cytometry analysis. Results are the mean ± SE from three independent experiments. Representative histograms of cell cycle analysis conducted with either NCI-H1703 (C) cells or A549 (D) cells treated with the indicated siRNAs. Symbols above a column indicate a statistical comparison of progression through each phase of the cell cycle by the indicated cells vs. the control cells transfected with NT siRNA (NS, not significant; *p < 0.05).
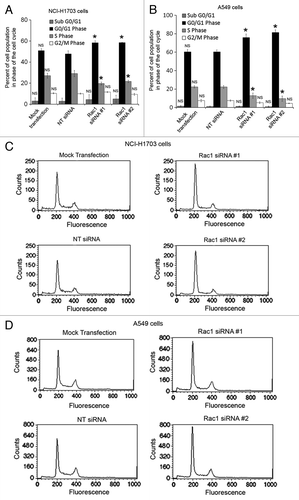
Figure 4. Silencing of Rac1 expression slows the migration of individual cells. (A–D) NCI-H1703 cells were transfected without siRNA (mock transfection) or with the indicated siRNAs, incubated for 48 h, and equal numbers of live cells were placed on colloidal gold coated with collagen Type I. Digital images of the cells were collected 24 h later. (E) The areas of the migration trails were measured after the cells migrated for 24 h, and the values were normalized to migration by the mock-transfected cells. Results are the mean ± SE from three independent experiments. Symbols above a column indicate a statistical comparison of migration by the indicated cells compared with control cells transfected with NT siRNA (NS, not significant; *p < 0.05).
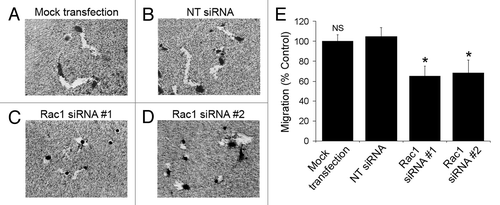
Figure 5. Pharmacological inhibition of Rac1 decreases proliferation and slows progression through the G1 phase of the cell cycle. (A) The effects of NSC23766 on Rac and Rho activity were measured by active Rac or Rho pull-down. Quantitative densitometry was conducted to determine the ratio of active GTPase to total GTPase detected in the immunoblots. The values are normalized to densitometric values obtained from ECL-Western blots of cells not treated with NSC23766. Results are the mean ± SE from three independent experiments. (B) Proliferation of NCI-H1703 cells was assayed by measuring [3H]thymidine uptake after a 24 h incubation with the indicated concentrations of the Rac inhibitor NSC23766. The values are normalized to [3H]thymidine uptake by cells treated without the drug. Results are the mean ± SE from three independent experiments conducted with six replicates for each treatment in each experiment. (C) The rate of proliferation was assessed by measuring [3H]thymidine uptake at the indicated times after incubation with NSC23766 for 24 h. (D) Changes in cell cycle progression were determined by incubation of NCI-H1703 cells with the Rac inhibitor NSC23766 (100 μg/ml) for 24, followed by flow cytometry analysis. (E) Histograms of cell cycle analysis conducted with NCI-H1703 cells treated with or without the Rac1 inhibitor NSC23766. Results are the mean ± SE from three independent experiments. Symbols above a column indicate a statistical comparison of cells treated with NSC23766 and control cells treated without NSC23766 (NS, not significant; *p < 0.01).
![Figure 5. Pharmacological inhibition of Rac1 decreases proliferation and slows progression through the G1 phase of the cell cycle. (A) The effects of NSC23766 on Rac and Rho activity were measured by active Rac or Rho pull-down. Quantitative densitometry was conducted to determine the ratio of active GTPase to total GTPase detected in the immunoblots. The values are normalized to densitometric values obtained from ECL-Western blots of cells not treated with NSC23766. Results are the mean ± SE from three independent experiments. (B) Proliferation of NCI-H1703 cells was assayed by measuring [3H]thymidine uptake after a 24 h incubation with the indicated concentrations of the Rac inhibitor NSC23766. The values are normalized to [3H]thymidine uptake by cells treated without the drug. Results are the mean ± SE from three independent experiments conducted with six replicates for each treatment in each experiment. (C) The rate of proliferation was assessed by measuring [3H]thymidine uptake at the indicated times after incubation with NSC23766 for 24 h. (D) Changes in cell cycle progression were determined by incubation of NCI-H1703 cells with the Rac inhibitor NSC23766 (100 μg/ml) for 24, followed by flow cytometry analysis. (E) Histograms of cell cycle analysis conducted with NCI-H1703 cells treated with or without the Rac1 inhibitor NSC23766. Results are the mean ± SE from three independent experiments. Symbols above a column indicate a statistical comparison of cells treated with NSC23766 and control cells treated without NSC23766 (NS, not significant; *p < 0.01).](/cms/asset/e18f411c-381a-47f8-880f-f99ecf66e3d1/kcbt_a_10920082_f0005.gif)
Figure 6. Inhibition of Rac1 reduces NF-κB transcriptional activity and treatment with BAY 11-7082 inhibits NF-κB activity and proliferation. (A) To examine NF-κB activity, NCI-H1703 cells were transfected without siRNA (mock transfection) or with the indicated siRNAs, cultured for 48 h, and then co-transfected with the pNifty-Luc NF-κB reporter plasmid and a β-galactosidase plasmid. After 24 h, the cells were incubated in the absence or presence of TNF-α (100 ng/ml) for 3 h, and luciferase activity was measured. Symbols above a column indicate a statistical comparison of the indicated treatment groups compared with control cells transfected with NT siRNA. (NS, not significant; *p < 0.05). (B and C) Cells were transfected with pNifty-Luc NF-κB reporter plasmid and a β-galactosidase plasmid and then incubated with the indicated concentrations of NSC23766 for 24 h or BAY 11-7082 for 12 h. Cells were then incubated in the absence or presence of TNF-α (100 ng/ml) for 3 h, and luciferase activity was measured. All NF-κB transcriptional activity results are the mean ± SE from three independent experiments normalized to β-galactosidase. (NS, not significant; *p < 0.01). (D) Cells were incubated with the indicated concentrations of BAY 11-7082 for 12 h and proliferation measured by [3H]thymidine uptake at the indicated time points. Symbols above a column indicate a statistical comparison of the indicated treatment groups compared with control cells transfected with NT siRNA or treated without drug (NS, not significant; *p < 0.001).
![Figure 6. Inhibition of Rac1 reduces NF-κB transcriptional activity and treatment with BAY 11-7082 inhibits NF-κB activity and proliferation. (A) To examine NF-κB activity, NCI-H1703 cells were transfected without siRNA (mock transfection) or with the indicated siRNAs, cultured for 48 h, and then co-transfected with the pNifty-Luc NF-κB reporter plasmid and a β-galactosidase plasmid. After 24 h, the cells were incubated in the absence or presence of TNF-α (100 ng/ml) for 3 h, and luciferase activity was measured. Symbols above a column indicate a statistical comparison of the indicated treatment groups compared with control cells transfected with NT siRNA. (NS, not significant; *p < 0.05). (B and C) Cells were transfected with pNifty-Luc NF-κB reporter plasmid and a β-galactosidase plasmid and then incubated with the indicated concentrations of NSC23766 for 24 h or BAY 11-7082 for 12 h. Cells were then incubated in the absence or presence of TNF-α (100 ng/ml) for 3 h, and luciferase activity was measured. All NF-κB transcriptional activity results are the mean ± SE from three independent experiments normalized to β-galactosidase. (NS, not significant; *p < 0.01). (D) Cells were incubated with the indicated concentrations of BAY 11-7082 for 12 h and proliferation measured by [3H]thymidine uptake at the indicated time points. Symbols above a column indicate a statistical comparison of the indicated treatment groups compared with control cells transfected with NT siRNA or treated without drug (NS, not significant; *p < 0.001).](/cms/asset/15463b73-ad6d-4504-8a9e-f01108bb924d/kcbt_a_10920082_f0006.gif)