Figures & data
Table 1. Characteristics of MPM patients and healthy volunteers
Figure 1. Flow cytometric analysis of iNKT cells and subsets from MPM patients and HV. PBMC were stained with anti-CD3 and anti-Vα24Vβ11 mAb. (A) Lymphocytes were gated on a side scatter (SSC)-forward scatter (FSC) dot plot. (B) In the lymphocyte gate, iNKT cells were identified as CD3+Vα24Vβ11+ double-positive cells. (C) Phenotypic analysis of iNKT cells was performed by CD4 and CD8 staining with anti-CD4 and anti-CD8 mAb. (D) A representative mean fluorescence intensity (MFI) histogram of CD3 expression on gate of lymphocytes. Representative flow cytometric dot plots of a HV (E) and a MPM patient (F); iNKT cell percentages are also shown in each panel. (G) The frequency of iNKT cells detected in the lymphocyte gate of MPM patients (n = 30; black bar) and HV (n = 22; unfilled bar). (H) Analysis of the frequency of iNKT cell subsets of MPM patients (black bars) and HV (unfilled bars). Results are expressed as mean ± SEM.
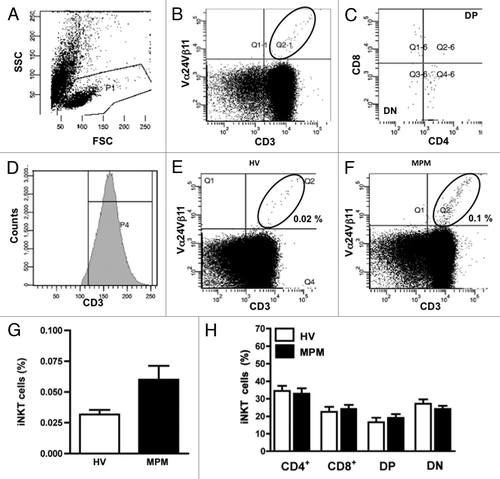
Figure 2. The frequency of circulating iNKT cells according to histological MPM subtypes. Scatter plot of iNKT frequencies in HV and biphasic- (B), epithelioid- (E) or sarcomatoid- (S) MPM patients. Horizontal bar is the median value for each group; *p < 0.05; **p < 0.01.
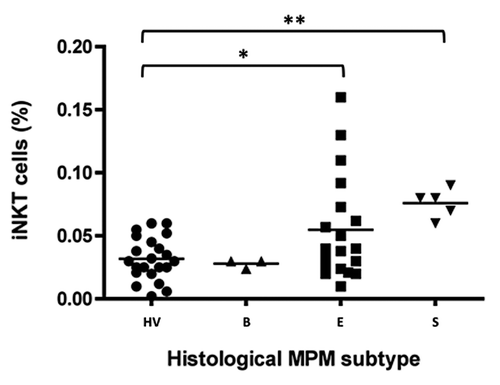
Figure 3. The frequency of iNKT cell subsets according to histological MPM subtypes. Analysis of the frequency of iNKT cell subsets was performed by CD4 and CD8 staining with anti-CD4 and anti-CD8 mAb. The values are expressed as percentage of iNKT cells expressing the molecules over the total iNKT cells. Data are the mean ± SEM; *p < 0.05.
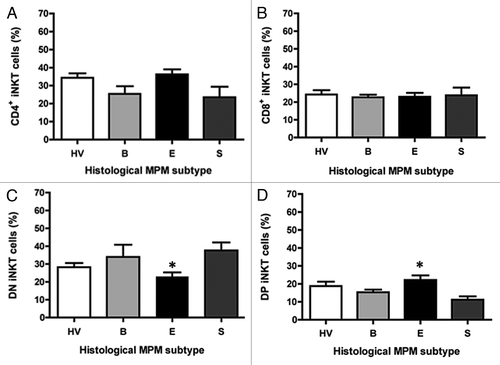
Figure 4. The frequency of iNKT cells and iNKT cell subsets in epithelioid and sarcomatoid MPM patients belonging to a certain clinical stage. Scatter plot of the iNKT cell frequency in HV and patients with epithelioid (A) or sarcomatoid (B) MPM at different clinical stages. Horizontal bar is the median value for each group. **p ≤ 0.01. (C and D) Analysis of the frequencies of iNKT cell subsets was performed by CD4 and CD8 staining with anti-CD4 and anti-CD8 mAb. The values are expressed as percentage of iNKT cells expressing the molecules over the total iNKT cells. Data are the mean ± SEM.
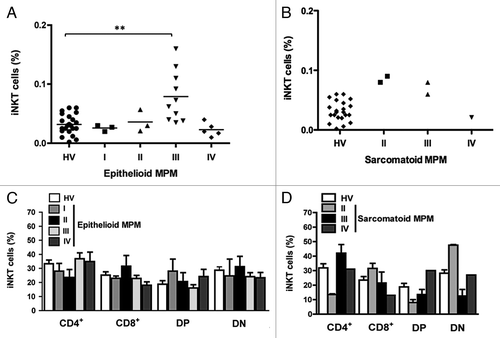
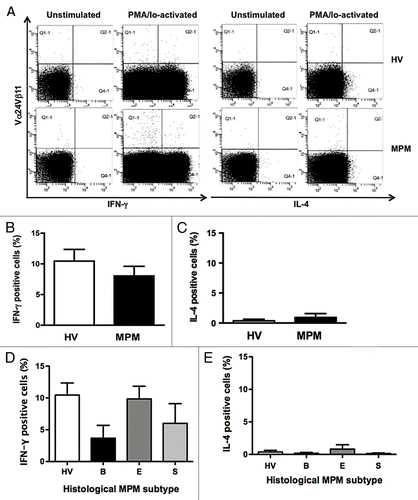
Figure 5. Intracellular cytokine expressions in iNKT cells. After stimulation of PBMC from HV and MPM patients with PMA/Io for 4 h, the cells were stained with anti-Vα24Vβ11 mAb followed by intracellular IFN-γ or IL-4 staining. (A) Representative dot plots displaying iNKT cell gates of unstimualted and PMA/Io-activated samples from a HV and a MPM patient. Percentages of IFN-γ and IL-4 positive cells in MPM patients (B and C) and in different MPM subtypes (D and E) compared with HV. Data are the mean ± SEM.