Figures & data
Table 1. Summary of genomic alterations in tumor samples
Table 2. Significantly enriched damaging mutations in the metastasis tumor sample
Figure 1. Structural variations indicated by Mutant (Mut) and Wild-type (WT) allele signal distributions on digital PCR array Y axis represents the ratio between the number of wells containing both mutant and wild-type allele signals (“Mut&WT”) to those containing only wild-type allele signals (“WT”), whereas X axis represents the ratio between the number of wells containing only mutant allele signals (“M”) over the wells with only wild-type allele signals (“WT”). The results from the germline heterozygous SNPs (red) show the distribution of the two ratios from the series of dilution of non-tumor samples (blood). The analysis of the KRAS variant shows a much higher ratio of the Mut&WT wells over the WT wells than what can be observed in the series of diluted non-tumor genomic DNAs, indicating that the high Mut&WT/WT ratio of the KRAS variant is unlikely due to the co-occupancy of the Mut and WT alleles within the same wells by chance. The ratio of the Mut wells than the WT wells is higher than the ratios observed for the germline heterozygous SNPs in the genomic DNAs of blood through the series of dilution, consistent with the estimated TP53 Mut allele frequency of more than 50%.
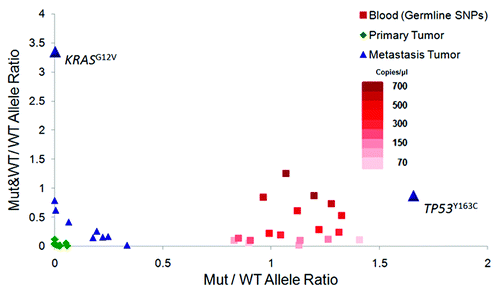
Figure 2. Most significant pathway involved the metastasis-enriched genes. Twelve genes harboring a total of 13 mutations were imputed into Ingenuity Pathway Analysis (IPA). Eight out of these 12 genes (in green) were clustered together into the same pathway, i.e., cancer (p value = 1.6 × 10−6 to 3.2 × 10−2) and tumor morphology (p value = 6.3 × 10−5 to 6.8 × 10−3). Molecules are represented as nodes and the biological relationship between two nodes is represented as an edge (line). Various shapes of the nodes represent the functional class of the gene product. Direct interactions are shown as solid lines whereas indirect as dashed lines.
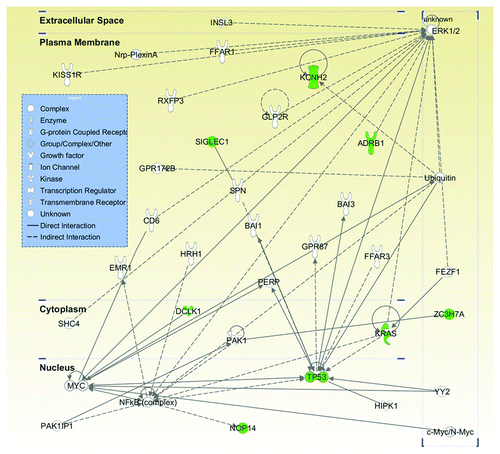
Figure 3. Effects of NOP14 expression on the migration and proliferation of pancreatic cancer cells. A, western-blotting showed the downregulated expression of NOP14 by siRNA and its effect on the expression of MAPK3/ERK1; B, comparisons of cell migration with/without knockdown of NOP14 in pancreatic cancer cell lines; C, comparisons of proliferation rates with/without knockdown of NOP14 in pancreatic cancer cell lines.
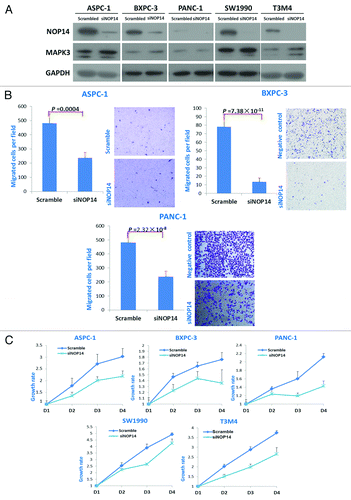
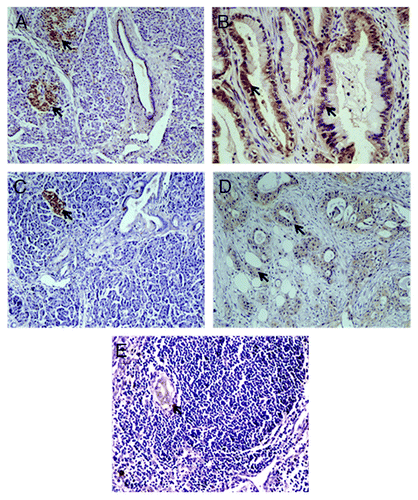
Figure 4. Immunohistochemistry staining of PDAC. A-B, IHC assays using antibody against KCNH2; arrows indicated expression of KCNH2 in pancreatic islet (A) and PDAC (B), but not in the normal pancreatic ductal and acinar tissues (A); C-E, IHC assays using antibody against SIGLEC1; arrows indicated expression of SIGLEC1 in pancreatic islet (C), PDAC (D) and metastasis lesion (E), but not in the normal pancreatic ductal and acinar tissue (C).