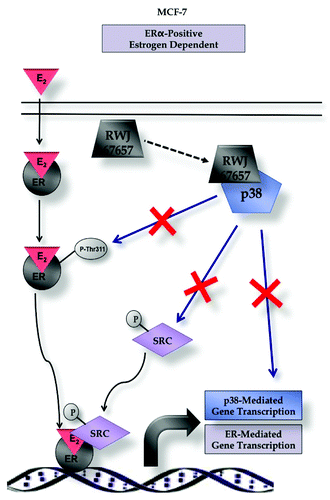
Figures & data
Figure 1. RWJ67657 inhibits p38-MAPK signaling. (A) MCF-7 cells were treated with E2 for the indicated times followed by western blot analysis. (B) MCF-7 cells were treated with RWJ67657, E2, tamoxifen (Tam) or ICI 182,780 (ICI) as indicated for 30 min followed by western blot analysis for total and phosphorylated levels of p38, hsp27 and MAPAPK. Blots shown are representative images of three separate experiments.
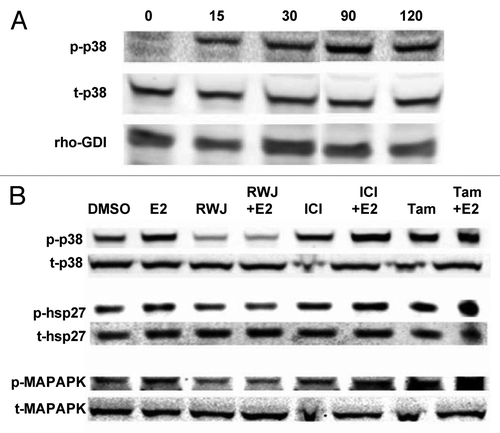
Figure 2. Inhibition of p38α decreases ERα transcriptional activity. (A) HEK293 or (B) MCF-7 cells were transiently transfected with pGl2-ERE2X-TK- luciferase plasmid. HEK293 cells were additionally transfected with pcDNA3.1B-ERα plasmid. Subsequently, cells were treated with veh (DMSO), E2 or RWJ67657. Cells treated with E2 were set to 1. Mean values ± SEM of three different experiments in triplicate are reported (***p < 0.001, **p < 0.01, *p < 0.05). (C) MCF-7 cells were transfected with an ERE-luciferase reporter and either empty vector or a dominant negative p38α construct (DN-p38α) followed by addition of either vehicle or 10 pM E2. Cells were assayed for luminescence and vector plus E2 was set to 100% for comparative purposes (*p < 0.05).
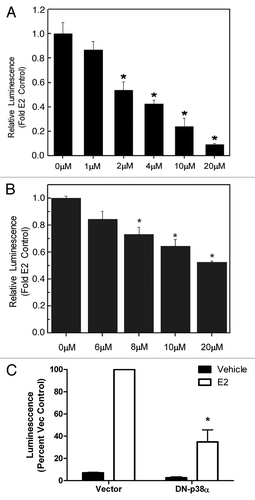
Figure 3. Effect of RWJ67657 on ER mutant function. HEK293 cells were transfected with ER mutant plasmids containing: (A) Thr311Ala phosphorylation site knockout, (B) AF-2 domain knockout or (C) AF-1 domain knockout. Subsequently, cells were treated as shown. Cells treated with E2 were set to 100. Mean values ± SEM of three different experiments in triplicate (*p < 0.05 compared with Vehicle control, #p < 0.05 compared with E2 control).
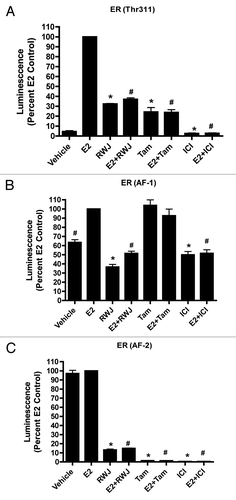
Figure 4. Effect of RWJ67657 on ERα (Tyr537Ser) and SRC Coactivation. (A) HEK293 cells were transfected with a constitutively active ERα mutant(Tyr537Ser) along with an ERE-luciferase construct. Subsequently, cells were treated with veh (DMSO), E2, RWJ67657 (RWJ), tamoxifen (Tam), ICI 182,780 (ICI) or a combination. Cells treated with E2 were set to 100. Mean values ± SEM of three different experiments in triplicate (*p < 0.05 compared with Vehicle control, #p < 0.05 compared with E2 control). (B) HEK293 cells were transiently transfected with a Gal-4-luciferase reporter construct and Gal-4-SRC-1, -2, -3 plasmids. Subsequently, cells were treated with DMSO (control) or increasing concentrations of RWJ67657 (RWJ). Cells treated with vehicle were set to 100. Mean values ± SEM of three different experiments in triplicate (*p < 0.05)
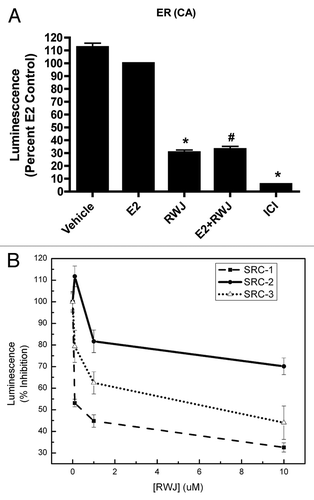
Figure 5. Pharmacological inhibition of p38α selectively targets breast cancer cell viability. (A) MCF-7 and (B) MCF-10A cells were treated with RWJ67657 for 24 h. Cell viability was determined using the MTT assay as described in the Materials and Methods section. Data are presented as percent viability compared with vehicle treated control cells. Mean values ± SEM of three different experiments in triplicate are reported (***p < 0.001, **p < 0.01, *p < 0.05).
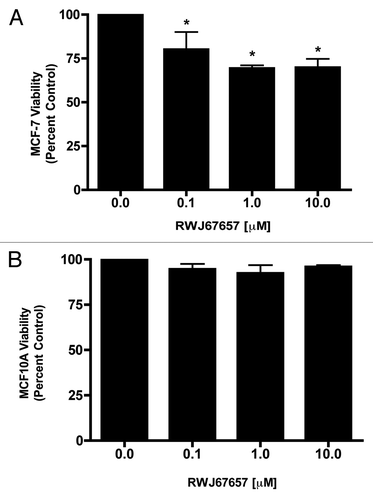
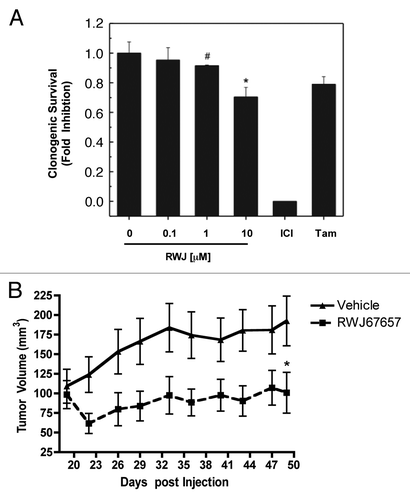
Figure 6. RWJ67657 inhibits breast cancer survival and tumorigenesis. (A) MCF-7 cells were plated at 500 cells per 60 mm2 dish. The following day, cells were treated with RWJ67657 for 10–14 d. Colonies ≥ 30 cells were scored as positive for colony formation. Data are presented as percent of vehicle treated samples. Mean values of ± SEM of three different experiments in triplicate are reported. (B) MCF-7 cells were injected in the mammary fat pads of female overiectomized mice with exogenous estrogen pellets. Tumors were allowed to form over 18 d. Mice were treated i.p with RWJ67657 for 41 d. Tumor volumes were measured every 3 d. Diminished volume of treatment tumors at endpoint were statistically significant from vehicle (*p < 0.05).