Figures & data
Figure 1. Gln dependency of NSCLC lines. Cells were grown in the presence or absence of Gln after which cell growth was assessed. Gln-dependency was rated arbitrarily: white bars represent Gln-dependent lines with growth in absence of Gln being equal or less than 30% of control; gray bars represent intermediate lines (growth between 30 and 70%), and black bars Gln-independent lines (growth equal or more than 70%).
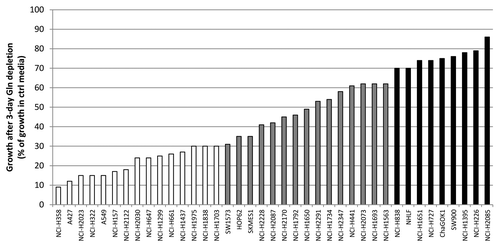
Figure 2. Amino acid analysis of NSCLC lines. (A) Relative free glutamine and glutamate levels in conditioned media. (B) Effect of Gln or Glc depletion of these lines on cell growth. Values were calculated from triplicate samples.
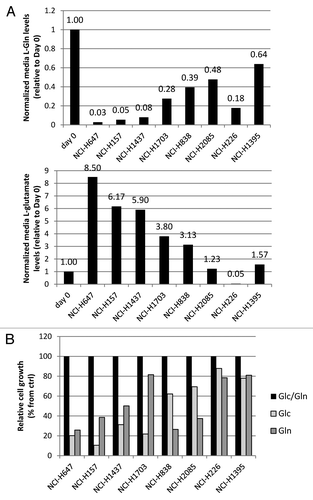
Figure 3. Knockdown of glutaminolysis targets in Gln-dependent and independent cell lines. Cell growth was assayed in NCI-H1703 (A) and NCI-H226 cells (B), after being transiently transfected with siRNA for a variety of enzymes involved in glutaminolysis, and subsequently grown in media with or without Glc for 4 d. The data points shown represent the mean ± SD of triplicate samples.
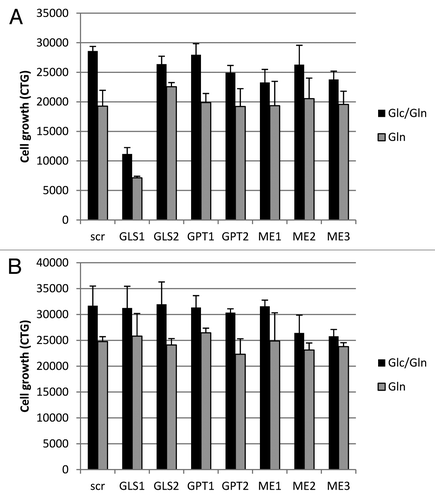
Figure 4. Importance of GLS1 function in Gln-dependent and independent lines. The data points shown represent the mean ± SD of triplicate samples. (A) Cells were grown for indicated times in the absence or presence of Gln, or treated with 10uM BPTES, after which cell growth was assessed. (B) Cells grown in the absence of Gln or treated with BPTES were co-treated with 2 mM or 5mM DM-aKG or DM-Glu for 2 d to rescue growth. Cell growth numbers were ATP values measured by CTG.
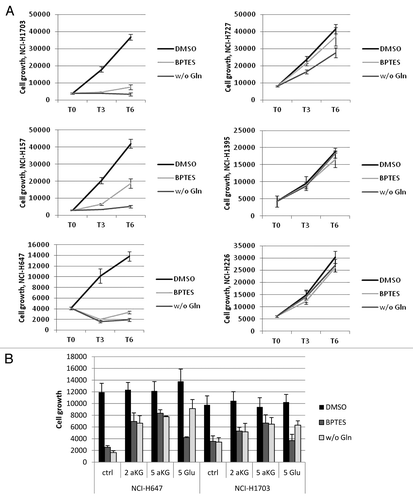
Figure 5. Analysis of GLS1 splice variant expression/ratio. (A) Schematic representation of GLS1 splice variant transcripts, and the location of the variant specific probes. (B) NSCLC cell lysates were analyzed for GLS1 expression. Transcript expression levels are depicted as Affymetrics MAS5 values. (C) Cell lines with varying GAC:KGA ratios were transiently transfected with siRNA oligos for either GAC or KGA after which GLS1 splice variant expression was analyzed by western blot analysis. (D) NSCLC tumors and matched normal lung tissues were analyzed for GAC:KGA ratio.
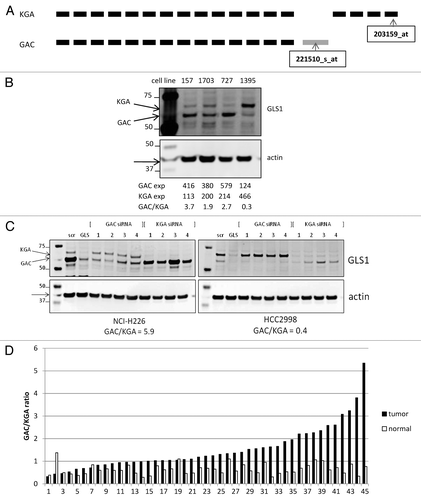
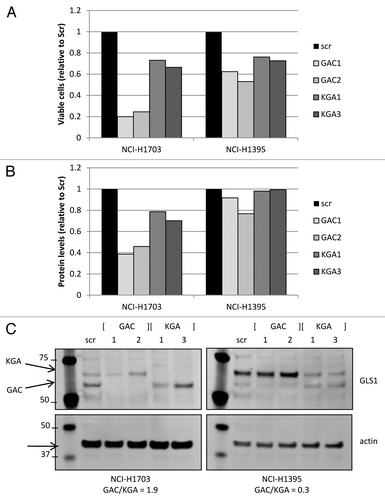
Figure 6. Transient GLS1 splice variant knockdown. Cell growth was assessed by measuring viable cell count (A) or total protein levels (B; BCA analysis) in cell lines that had GLS1 splice variant transiently knocked down by specific siRNA oligos directed against either GAC or KGA. Equalized protein lysates of experimental samples (from A) were analyzed for GLS1 splice variant and actin expression (C).