Figures & data
Table 1. Evaluation of DHFR activity was accomplished by measuring absorbance of NADPH using spectrophotometric analysis at 340 nm
Figure 1. Three thousand CCRF-CEMR or CCRF-CEMS were plated in RPMI media containing varying concentrations of drug, highest concentration of 20 μM, and incubated at 37ºC for 96 h. Cytotoxicity was measured using the trypan blue exclusion assay and cell viability was evaluated using the Vi-CELL Series Cell Viability Analyzer.
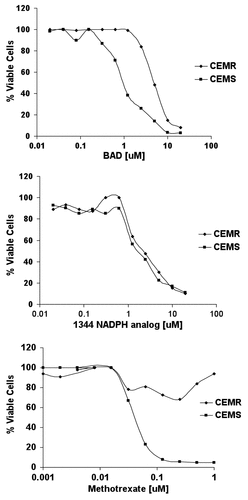
Figure 2. In order to evaluate the potential for BAD to act as a substrate of phosphorylation via human recombinant NAD kinase, 10 μM BAD was incubated for 1 h in a solution containing 50 mM Tris/HCL, pH 8.0, 20 mM MgCl2, 5 mM ATP, and 1–4 μg of human recombinant NAD kinase. To validate the activity of NAD kinase, 10 μM NAD was also incubated in the aforementioned solution for 1 h. Following incubation, samples were evaluated via HP LC using a Prtisil-10 SA X column at an absorbance of 254 nm as described by Saunders et al.Citation14 Data shows samples in which NAD was incubated with reaction solution lacking NAD kinase (A), NAD incubated in the reaction solution with 2 μg NAD kinase (B), BAD incubated with reaction solution lacking NAD kinase (C), 10 μM BAD standard (D), and 10 μM BAD incubated in reaction solution containing 4 μg NAD kinase (E).
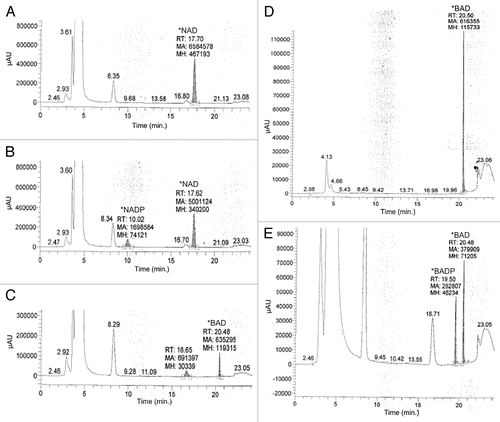
Figure 3. Sixty million CCRF-CEMR cells were plated in RPMI media with or without 50 μM BR and incubated at 37ºC for 3 h. Following incubation, cells were lysed with 140 μL of ice cold HCl04, kept at room temperature for 10 min and then spun down for 1 min at 16,000 RPM. An amount of 112 μL of ice cold K2CO3 was added to each sample, and samples were then kept at -20ºC for 15 min. The samples were centrifuged and the supernatant was collected and analyzed via HP LC at conditions described by Saunders et al.Citation14 (A) HP LC data of cell lysate from those cells not treated with BR, and (B) shows HP LC data of cells lysate from those cells treated with 50 μM BR. Finally, (C) is the HP LC data of 50 μM BR in water.
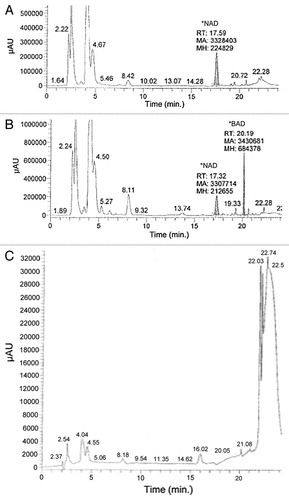
Figure 4. Five million CCRF-CEMR cells were plated in RPMI media containing 10 μM BR, BAD or no drug. Cells were allowed to incubate for up to 24 h harvested and then analyzed for NADP+/NADPH levels as described in Methods. (A) Levels of NADPH for the three experimental groups at 0, 8 and 24 h. (B) Levels of NADP+ at the same conditions. 132, BR; 106, BAD.
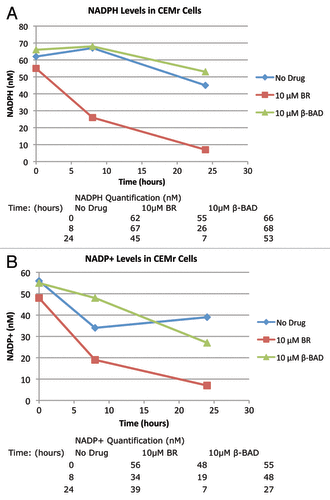
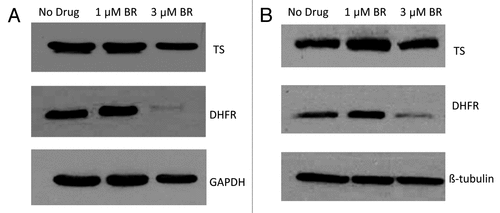
Figure 5. The effect of BR treatment on DHFR levels. CCRF-CEM/S cells were pelleted in RPMI media with BR (1 or 10 μM ) for 24 h (A) or 48 h (B). Following treatment cells were lysed and protein levels analysed by western blotting.