Figures & data
Table 1. Clinical characteristics of patients
Figure 1. Viability of CLL cells (A), differential scanning calorimetry profiles of nuclear fraction (B) and changes in expression of apoptosis-related proteins (Mcl-1, Bcl-2, procaspase-3, Noxa) in PBMC lysates (C), isolated from blood of patients PZ and RJ, incubated for 48 h with and without RCM combination. Immunoblot analysis of PBMCs after their exposure to drug combination were analyzed by alkaline phosphatase method in the presence of appropriate antisera. Protein lysates (50 μg) were loaded into 12.5% polyacrylamide gels; β-actin was used as loading control; Cpex - excess heat capacity.
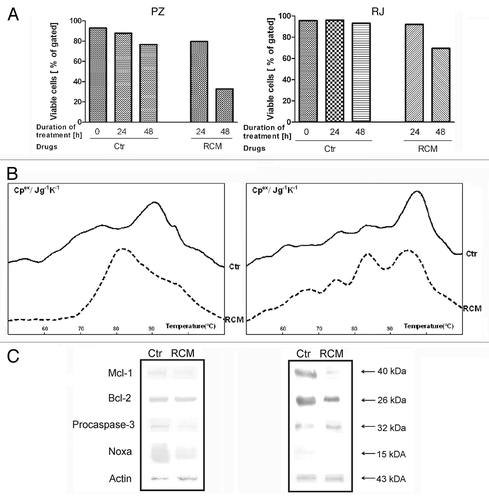
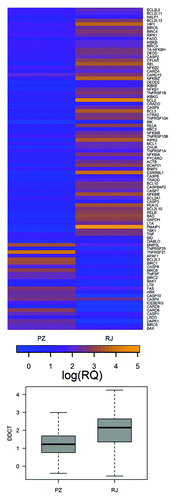
Figure 2. Microarray analysis indicating changes in expression of 89 apoptosis-related genes in two CLL samples following immunotherapy (top); boxplots of the DDCT values of samples from patients PZ and RJ (bottom). The rows and the columns represent individual genes and mRNA samples, respectively. The relative level of gene expression is depicted according to the color scale shown below the matrix. The accuracy of reading in the presented experiments is illustrated by boxplots of the signal (signal DDCT ~-logRQ).