Figures & data
Figure 1. AZ58 overcomes resistance to gemcitabine and cisplatin in select urothelial cancer cell lines. Ten urothelial cancer cell lines were exposed to 1 μM each of gemcitabine and cisplatin for 48 h. Cells were harvested for PI-FACS analysis. Asterisks indicate cell lines sensitive to the drug combination (30% DNA fragmentation). Error bars = standard error. The same 10 urothelial cancer cell lines were exposed to AZ58 at 30 nM in addition to gemcitabine and cisplatin. Double asterisks indicate cell lines sensitive to the drug combination (30% DNA fragmentation).
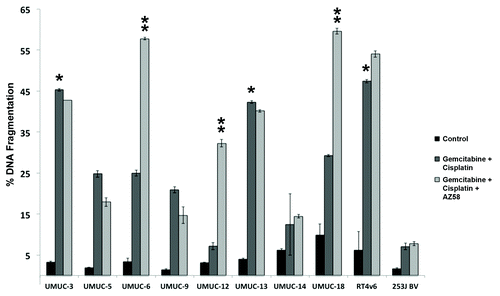
Figure 2. Constitutive protein expression is not predictive of drug (chemotherapy and/or Smac mimetic) sensitivity. Western blotting analysis of the ten urothelial cancer cell lines for baseline expression of IAP family, BCL family and Smac protein.
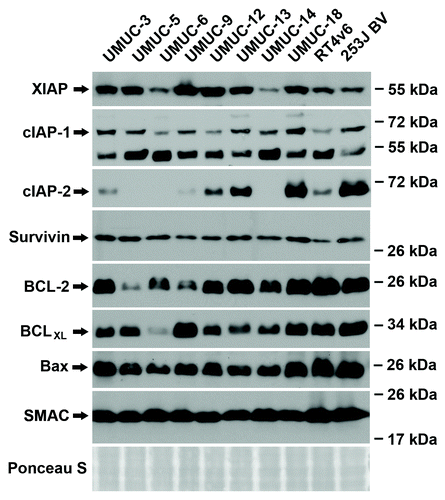
Figure 3. AZ58 potentiates cell death through apoptosis. (A) Western blotting of the urothelial cancer cell lines considered resistant to gemcitabine and cisplatin (UMUC-12, UMUC-6, and UMUC-9). Treatment conditions are as labeled. (B) Image J quantitation of caspase cleavage products. Bar graphs demonstrate relative densities compared with the control. (C) Corresponding graphs of DNA fragmentation from PI-FACS are demonstrated.
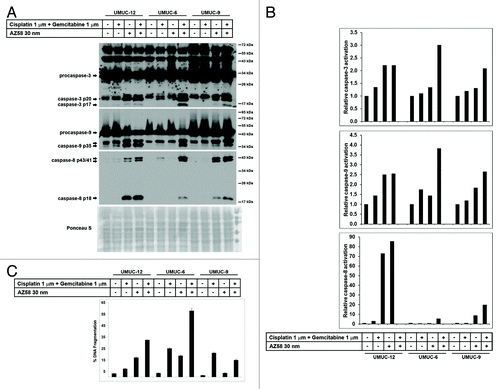
Figure 4. Degradation of cIAP-1 corresponds to cell death conditions in urothelial cancer cell lines identified as resistant to gemcitabine and cisplatin. Western blotting of urothelial cancer cell lines resistant to gemcitabine and cisplatin were subjected to various conditions as indicated. Antibodies for selected members of the IAP family, BCL family, and Smac were immunoblotted.
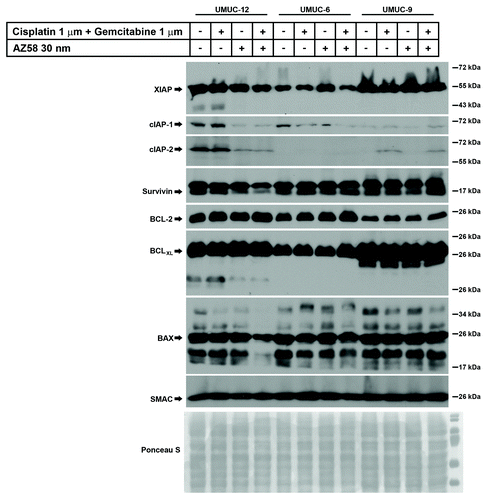
Figure 5. AZ58 treatment results in reduced tumor burden; however, in a non-statistically significant fashion. (A) Microscopy of UMUC-6 cells demonstrating differential cell death in the various treatment conditions as labeled. Corresponding images show resultant graphs of PI-FACS demonstrating DNA fragmentation. (B) Female athymic nude mice were injected with UMUC-6 cells and treated as described in Materials and Methods. Tumor volumes were calculated and plotted. n = 6 mice for each condition. Error bars = standard error. (C) TUNEL staining of fragmented apoptotic DNA/nuclei to detect the efficacy of AZ58 and chemotherapy in vivo (green) and the total nuclei stained using propidium iodide (Red). Magnification 10 ×. (D) Mean number of TUNEL positive nuclei from 10 representative images.
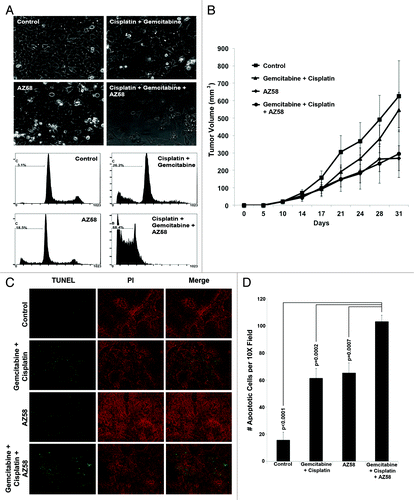
Figure 6. AZ58, in combination with chemotherapy, functions through decreased cellular proliferation and decreased microvessel density. (A) Immunohistochemistry with Ki-67 is displayed. Imaging was performed at 20 × magnification. Representative images are shown. (B) Positive Ki-67 nuclei were counted, averaged, and plotted on a bar graph. Error bars represent standard error. P values were calculated compared with the control condition. n = 10 images per treatment condition. (C) Immunohistochemistry using an antibody for CD-31 is displayed. Imaging was performed at 20 × magnification. Representative images are shown. (D) Positive foci of CD-31 were counted, averaged, and plotted on a bar graph. Error bars represent standard error. P values were calculated compared with the combination condition. n = 5 per treatment condition.
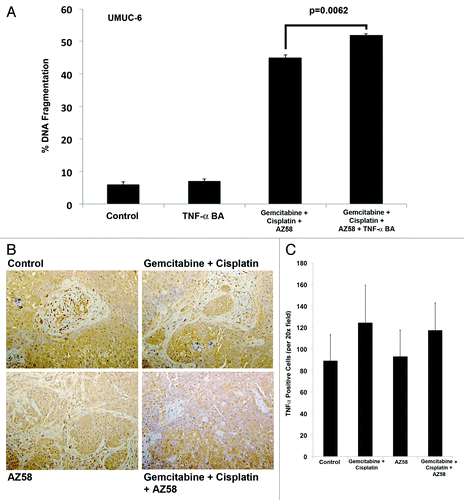
Figure 7. Cell death in UMUC-6 from AZ58 is mediated through a TNF-α-independent mechanism. (A) The effects on UMUC-6 of chemotherapy and AZ58 were not mitigated by the TNF-α blocking antibody. (B) Representative images of IHC for TNF-α demonstrate no difference in expression between treatment conditions. (C) Ennumeration with cell counting confirms the lack of a statistical difference in TNF-α expression.