Figures & data
Figure 1. CD31-MicroBeads purification of endothelial cells from ovarian carcinoma tissue. (A) Morphology and phenotype of EOT microvessels. (a and b)Tissue staining reveals CD31+ and endoglin+ vasculature. (B) The purity of endothelial cells was monitored by FACS analysis of the CD31 surface antigen before and after magnetic sorting (n = 3). (C) Representative flow cytometric analysis of CD31+ cells showing the expression of the endothelial markers CD146 and endoglin. ODMECs are negative for a-SMA and cytokeratin-20.
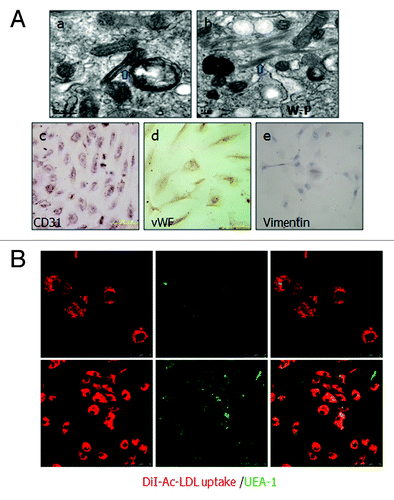
Figure 2. Identification of cultured human ovarian carcinoma endothelial cells. (A) (a and b) ODMECs are positive typical markers, including CD31 and vWF. (c) ODMECs are negative for vimentin. (B)Fluorescence microscopy showed a bright homogenous fluorescence of all cells with UEA-I lectin (green), in contrast to the more restricted granular immunofluorescence with the internalized fluorochrome-conjugated Dil-Ac-LDL (red).
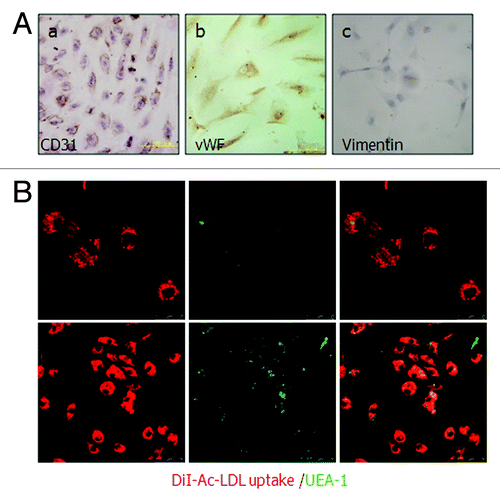
Figure 3. Culture characteristics of ODMECs. (A) Morphology of ODMECs. (a) Individual cells show formation of lamellipodia and filopodia in the first ODMECs passage. (b) The morphology of primary and P1 ODMECs was variable with respect to size and shape. (c) At high passages ODMECs showed regular morphology but increased in size and replicative senescence.(B)Electron microscopy illustrates (a) Weibel–Palade bodies, (b) numerous microtubules and microfilaments. (c) Tight junctions between cells (purple arrow). (C and D) ODMECs showed overexpression of growth factor receptors endoglin and VEGFR-2.
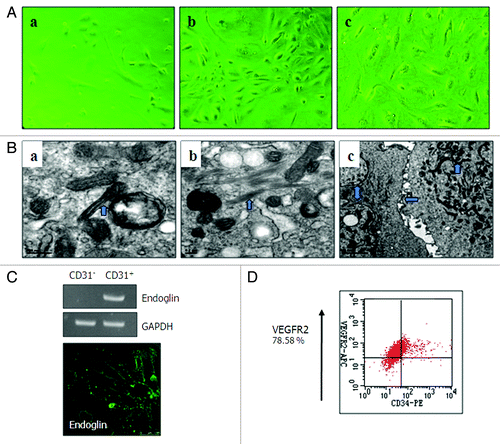
Figure 4. (A) ODMECs exhibit angiogenic capabilities on fibrin gels at the indicated plating density. (a) Cells initially presented as a single cell sprouting. (b) After extended time in culture, the sprouting cells interconnected and grew in tube-like structures. (c) Networks of cords then formed. (B)Endoglin was expressed at higher levels during the tubular structure formation stage (7 d) as opposed to the stage of stable-formed networks (14 d) (n = 3, *P < 0.05).
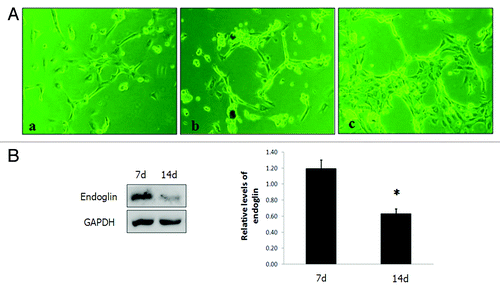
Figure 5. Endoglin RNA interference in ODMECs. (A) Quantification of transfection efficiency in ODMECs for GFP using lentiviral vector transduction by both fluorescence microscopy and flow cytometry. (B) RT-PCR analysis reveals a significant reduction (n = 3, P < 0.05) in endoglin transcript levels following transduction of LV-END in ODMECs. Each band was quantitated against the loading control GAPDH: ODMECs alone (1.02 ± 0.06), siRNA control LV-H (1.07 ± 0.08) and LV-END (0.54 ± 0.04). (C) Western blot analysis showed that LV-END (0.17 ± 0.08) significantly decreased endoglin protein expression compared with LV-H (0.95 ± 0.33) and non-transfected ODMECs (0.76 ± 0.08) (MOI = 50,48 h) (n = 3, P < 0.05).
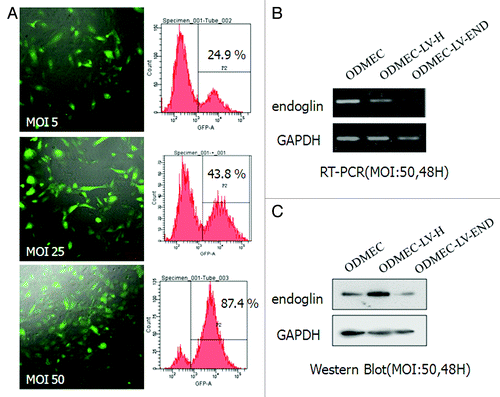
Figure 6. Proliferation and migration of ODMECs transfected with LV-END and LV-H siRNA. (A) A wound-healing assay demonstrates that the motility of ODMECs were incubated for 16 h after administration of the scratch, which were not statistically different. Scale bar; 50 μm. (n = 3; *P > 0.05). (B) A cell proliferation assay demonstrates that LV-H-ODMECs grow faster than LV-END-ODMECs at 3 to 6 d post-infection, Meanwhile, the doubling time for ODMECs inhibited (72–120 h). Cells could not enter the logarithmic phase of growth. (n = 3; *P < 0.05, **P < 0.001).
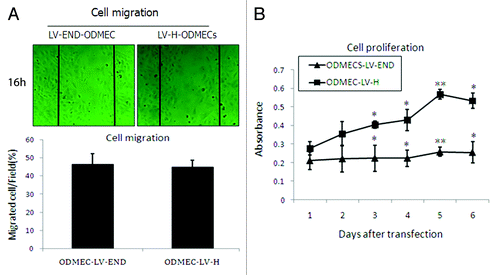
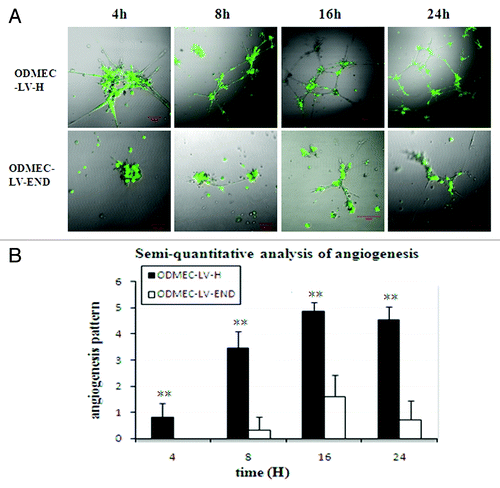
Figure 7. Endoglin-expressing ODMECs form vascular tubes more efficiently.(A) ODMECs vascular tubule formation was assessed on Matrigel at 4, 8, 16, and 24 h in LV-H and LV-END transfected ODMECs.(B) Vascular tube structures were quantitated using semi-quantitative analysis (n = 5; **P < 0.001).