Figures & data
Figure 1. NUSAP1 is required for high levels of DSB repair by homologous recombination. (A) HeLa-DR cells, which contain integrated in the genome a recombination substrate for assaying HDR, were transfected with the indicated siRNAs, and then 2 d later transfected with the same siRNA plus a plasmid for the expression of the I-SceI endonuclease. Results were obtained in triplicate and normalized relative to the control (Con) siRNA with SEM. BRCA1 (BR) and NUSAP1 (NUS) siRNAs are indicated. A two-tailed Student t test was done for the NUSAP1 siRNAs compared with the control siRNA; P values for comparisons to the control siRNA were: 0.004195 for BRCA1; 0.0003138 for NUS-si2; 0.002255 for NUS-si3; and 0.002923 for NUS-si4. (B) Whole cell lysates were prepared from the leftover cells after assaying for HDR activity and analyzed by immunoblotting for antibody specific for NUSAP1 (top) and GAPDH as loading control (bottom). (C) HDR assay was done for HeLa-DR cells that were not transfected with I-SceI expressing plasmid (lane 1) or transfected with the following siRNAs and plasmids along with the I-SceI expressing plasmid: control siRNA and empty plasmid vector (lane 2); NUS-si2 and empty plasmid vector (lane 3); and NUS-si2 plus a plasmid encoding NUSAP1 with a conservative point mutation that renders the mRNA resistant to the siRNA (Res, lane 4). A two-tailed Student t test was done for NUSAP1 siRNA vs. control siRNA (lane 3 vs. lane 2; P = 0.02289) and NUS-si2 plus rescue transfection vs. NUS-si2 plus vector only (lane 4 vs. lane 3; P = 0.023224). (D) Samples from (C) were analyzed by immunoblotting with antibody specific to NUSAP1 (top) and RNA helicase A (RHA, bottom) as loading control. (E) HeLa-SA cells, which contain integrated in the genome a substrate that specifically measures recombination via the SSA pathway, were transfected with the indicated siRNAs. Results, in triplicate, were normalized to the control siRNA. A two-tailed Student t test was done comparing to the control siRNA; P values were: 0.006797 for BRCA1; 0.01224 for NUS-si2; and 0.02585 for NUS-si3.
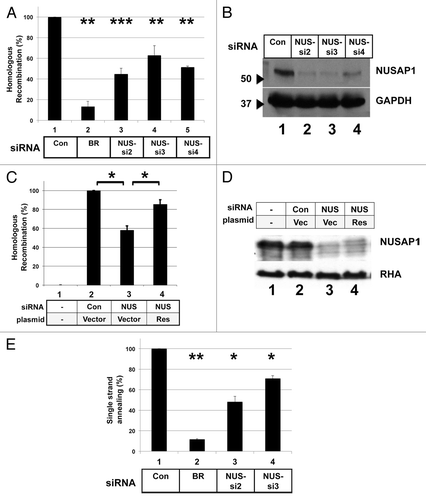
Figure 2. Depletion of NUSAP1 from Hs578T breast cancer cells results in supernumerary centrosomes. Hs578T cells were transfected with the indicated siRNA and a plasmid that expressed GFP-centrin2.Citation47 Centrosomes were counted in over 100 cells in each sample. Results, in triplicate, were normalized to the control siRNA.
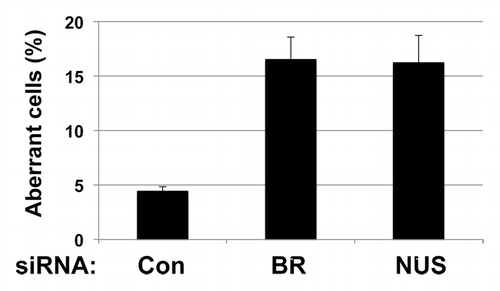
Figure 3. NUSAP1 protein expression occurs in S/G2 phases of the cell cycle. HeLa cells were double-thymidine blocked in early S phase and released for the indicated times (lanes 1–4) or thymidine-nocodazole blocked in mitosis and released for the indicated times (lanes 5–8). Immunoblots measure the indicated protein abundance for BRCA1, NUSAP1, cyclin A, and cyclin B1 with matched loading controls, as indicated.
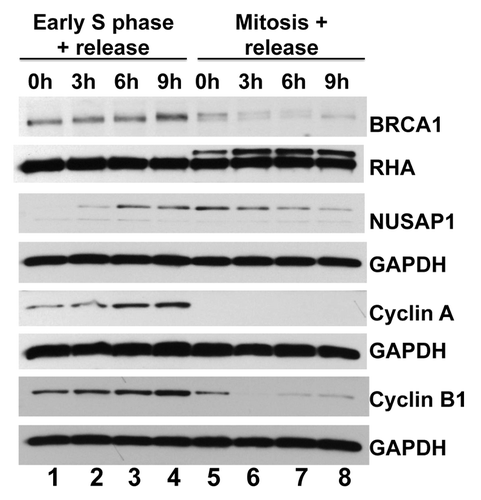
Figure 4. Expression of BRCA1 from a plasmid suppresses effects of NUSAP1 depletion. (A) HeLa-DR cells were transfected with the indicated siRNAs and a plasmid for the expression of BRCA1 (lanes 2 and 4). Results were obtained in triplicate and normalized relative to the control siRNA. A two-tailed Student t test compared the effects of NUSAP1 si-2 to the control siRNA (lane 3 vs. lane 1; P = 0.001603) and the effect of rescue with BRCA1-expressing plasmid (lane 4 vs. lane 3; P = 0.04048). (B) Hs578T cells were transfected with the indicated siRNAs and plasmids, along with an expression plasmid for GFP-centrin2, and the percentage of cells with supernumerary centrosomes is indicated. Results were obtained in triplicate plus SEM. A two-tailed Student t test compared the effects of NUSAP1 si-2 to the control siRNA (lane 3 vs. lane 1; P = 0.04099) and the effect of rescue with BRCA1-expressing plasmid (lane 4 vs. lane 3; P = 0.04859).
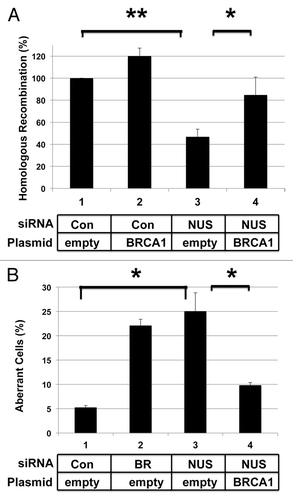
Figure 5. Depletion of NUSAP1 affects BRCA1 protein but not mRNA levels. (A) Two days following transfection of HeLa cells with the indicated siRNA, RNA was isolated and quantified by qPCR. Results, in triplicate, from BRCA1-specific primers are indicated in gray, and results from NUSAP1-specific primers are indicated in black. Results were normalized to the cells transfected with the control siRNA and presented with the standard error of the mean. (B) Cells were transfected with control (C; lanes 1, 4, 7, 10), NUSAP1 (N; lanes 2, 5, 8, 11), and BRCA1 (B; lanes 3, 6, 9, 12) specific siRNAs. Following double thymidine block and release for the indicated times, cell lysates were prepared and analyzed with the indicated immunoblots. RHA and GAPDH were loading controls. (C) Cells were transfected with control siRNA (C; lanes 1, 3) or NUSAP1 siRNA2 (N; lanes 2, 4) and subjected to a double thymidine block as in panel B. Two hours after thymidine release, 20 µm MG132 was included in media for another 4 h (lanes 3, 4); DMSO vehicle was included in media at the same time in lanes 1 and 2.
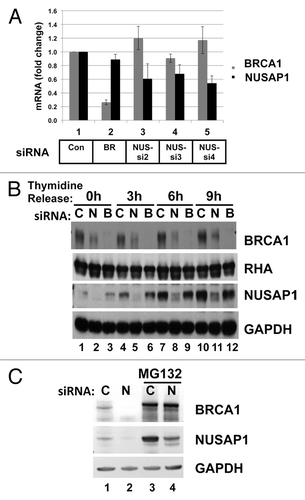
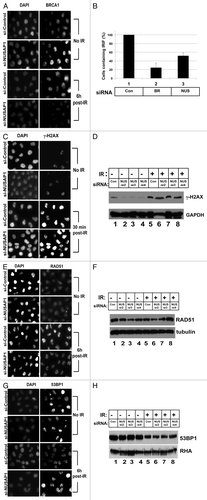
Figure 6. NUSAP1 depletion affects BRCA1 and RAD51 localization to DNA damage induced foci. (A) HeLa cells were transfected with control or NUSAP1 specific siRNAs and stained for DNA content (left) or BRCA1 content (right) before (top) or after (bottom) 10 Gy of ionizing radiation. (B) The number of cells with detectable BRCA1 IRIF from (A) is indicated, normalized relative to the control siRNA plus the SEM (C) Cells transfected as in panel A were stained for γ-H2AX, as indicated. (D) Cell lysates were prepared from the transfected cells in (C) and immunoblots were probed with antibody specific to γ-H2AX, and GAPDH is the loading control. (E) Cells transfected as in (A) were stained for RAD51, as indicated. (F) Cell lysates were prepared from the transfected cells in (C) and immunoblots were probed with antibody specific to RAD51, and tubulin is the loading control. (G) Cells transfected as in (A) were stained for 53BP1, as indicated. (H) Cell lysates were prepared from the transfected cells in (C) and immunoblots were probed with antibody specific to 53BP151, and RHA is the loading control.