Figures & data
Figure 1. Localization of p53 in untreated and drug treated MCF-7 breast cancer cells. MCF-7 cells were treated with doxorubicin or 5-fluorouracil for 4–12 h or untreated (UT), then nuclear and cytoplasmic extracts were produced as described in the Materials and Methods section. Protein from each extract (25 μg) was loaded onto a 10% polyacrylamide gel, transferred to nitrocellulose and then probed for either p53 or β-actin. Western blot analysis shows the distribution of p53 in nuclear vs. cytoplasmic fractions for MCF-7 untreated, doxorubicin, and 5-fluorouracil treated cells for various times. β-actin was used as a loading control. The p53 protein increases dramatically after drug treatment and is nearly exclusively located in the nucleus of MCF-7 breast cancer cells in all conditions.
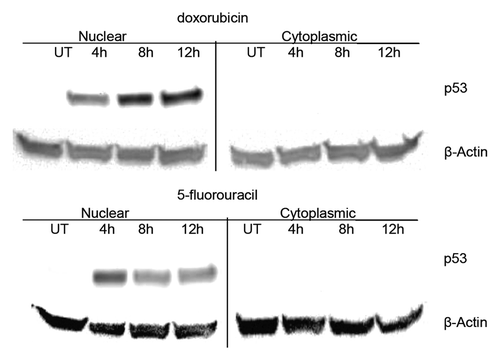
Figure 2. Electrophoretic mobility shift assay (EMSA) using p53 from MCF-7 nuclear extracts and p53 target gene response elements (REs). MCF-7 cells were treated with doxorubicin (dox) or 5-fluorouracil (5FU) for 4–12 h or untreated (UT), then nuclear extracts were produced as described in the Materials and Methods section. DNA binding was performed by EMSA as described by Chandrachud and GalCitation57 and Ray and colleaguesCitation51 using the fluorescently labeled p21 sequence. (A and C) Aliquots of MCF-7 untreated and drug-treated nuclear extracts representing 20 μg total protein were reacted with 5 pmoles of 6-FAM p21 gene RE (A) or the p53 REs from the puma, mdm2, bax, and survivin genes (C). A shifted band was observed for each reaction with a commensurate change in the unbound DNA. The standard deviation of the mean relative to untreated control for each sample was calculated at a 95% confidence level for 3 replicates for the bound DNA sequence. The average change in band density of the shifted band (protein + DNA) was found to be insignificant in each case (avg. P = 0.15). (B and D) Aliquots of MCF-7 untreated (UT) and drug-treated nuclear extracts representing 40 pg of p53 were reacted with 5 pmoles of 6-FAM p21 gene RE (B) or the p53 REs from the puma, mdm2, bax, and survivin genes (D). A shifted band was observed for the reaction with the untreated sample with a commensurate decrease in the unbound DNA compared with the reaction without extract. However, there is minimal detection of a shifted band after drug treatment (either dox or 5FU), as well as an increase in the unbound DNA signal. The standard deviation of the mean relative to untreated control for each sample was calculated at a 95% confidence level for 3 replicates for each bound DNA sequence. The average change in band density relative to the shifted band (protein + DNA) was found to be significant in each case (avg. P = 0.006). DNA only (p21) and extract only (Ext) were used as controls (A and B). Unexpectedly, the increase in level of p53 is associated with a decrease in DNA binding.
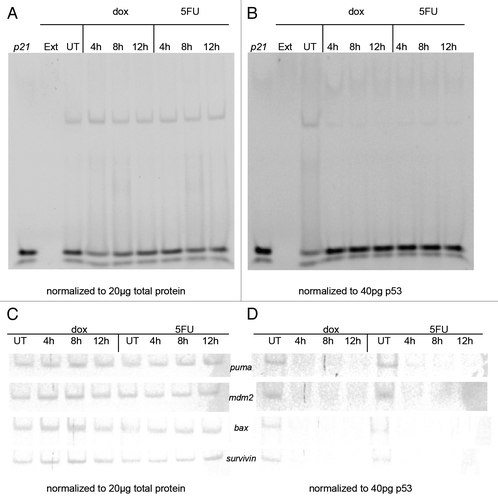
Figure 3. Chromatin immunoprecipitation (ChIP) of p53 from MCF-7 cells following treatment with 5-fluorouracil and doxorubicin. MCF-7 cells were treated with doxorubicin or 5-fluorouracil for 4–12 h or untreated, then chromatin was produced as described in the Materials and Methods section. ChIP was performed using anti-p53 antibodies then PCR done on the isolated chromatin using the p21, mdm2, and bax gene RE sequences as described. The raw density values detected in gels (data not shown) were obtained and plotted as compared with that detected with an untreated sample. The standard deviation of the mean relative to input of each sample, shown as the error bars, was calculated at a 95% confidence level for 3 replicates. (A) After a 12 h treatment with doxorubicin, there is a significant increase in the association of p53 with p21 (P = 0.0006) and mdm2 (P = 0.0003). (B) There is a significant increase in the association of p53 with p21 (P < 0.0001) and mdm2 (P = 0.0003) after a 12 h treatment with 5-fluorouracil. The bax gene was not analyzed at intermediate times, only with untreated and 12 h treated samples.
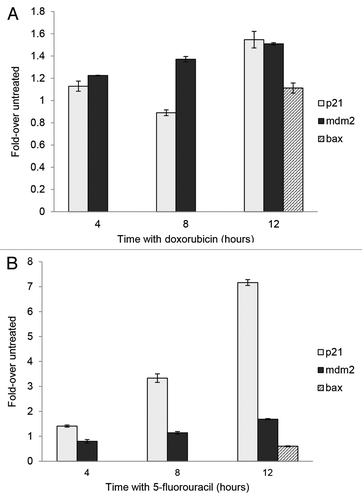
Figure 4. Whole cell extracts from MCF-7 cells treated with drugs and p53 siRNA. (A) MCF-7 cells were treated with 5-fluorouracil (5FU) or doxorubicin (dox) for 12 h or with UV light (UV) then left for 24 h or left untreated (UT). (B) MCF-7 cells were pretreated with 5 nM p53-specific or non-targeting siRNA for 24 h, followed by treatment with drugs or UV as above. Whole cell extracts were prepared using RIPA buffer and 30 μg total protein separated on either 10% polyacrylamide Tris-glycine SDS gels for blots probed for p53, Mdm2, and MdmX or 16.5% polyacrylamide Tris-tricine SDS gels for blots probed for p21 as described in the Materials and Methods section. The proteins were transferred to nitrocellulose and then probed for the indicated proteins as described in the Materials and Methods section. The results indicate that the p53 protein is required for the accumulation of both Mdm2 and p21 following drug treatment but not for changes in the MdmX protein.
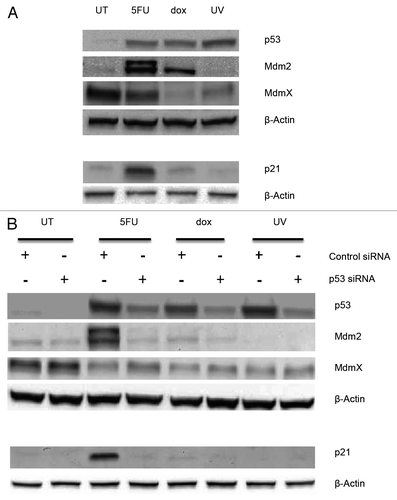
Figure 5. Detection of serine 15 phosphorylation in p53 from MCF-7 treated cells MCF-7 whole cell extracts from cells treated with doxorubicin (dox) or 5-fluorouracil (5FU) for 12 or 24 h, or treated with UV followed by incubation for 12 or 24 h, were isolated and used. Extracts containing ~120 μg total protein were separated using a polyacrylamide gel, transferred to a nitrocellulose membrane, and the membrane probed with Phosphodetect™ anti-p53 pSer15. We detect phosphorylation at serine 15 in both UV and dox treated extracts after 12 h, but do not detect the modification in 5FU treated extracts after 24 h. We estimate >6000 pg of p53 was loaded from the dox and 5FU treated extracts after 24 h of treatment, while the amount of p53 in UV treated extracts is estimated to be ~900 pg p53 based on ELISA. The p53 detection is not shown from this gel as it would overwhelm the detection.
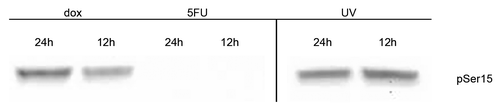
Figure 6. Detection of enzymes potentially responsible for phosphorylation of p53 at serine 15. Nuclear extracts containing 25 μg of total protein from MCF-7 cells treated with doxorubicin or 5-fluorouracil for 4–12 h, 24 h after UV treatment, or untreated (UT) were separated using a 10% polyacrylamide gel, the proteins transferred to nitrocellulose, and the membranes probed with ATM or Chk2, with anti-phospho-ATM pSer1981 or anti-phospho-Chk2 pThr68 antibodies. The blots were subsequently probed for β-actin. ATM and Chk2 proteins are detected in drug- and UV-treated extracts, but the presence of phosphorylated ATM (pSer1981) was only detected in doxorubicin- and UV-treated extracts. Phosphorylated Chk2 (pThr68) was detected only in UV-treated extracts. The ATM kinase, but not Chk2, was detected in untreated cell (UT) extracts.
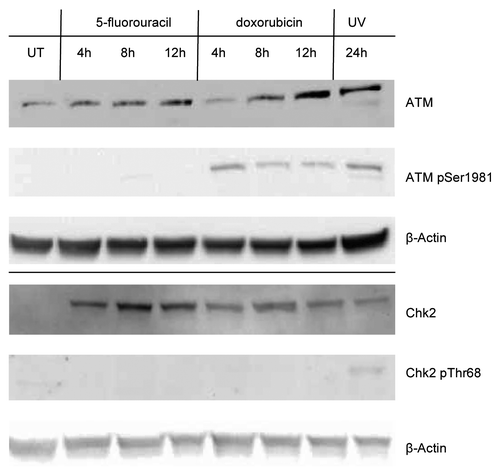
Figure 7. Whole cell extracts from MCF-7 cells treated with MG132 Cells were first treated with 5-fluorouracil (5FU) or doxorubicin (dox) for 6 h or with UV light (UV) then left for 18 h or left untreated (UT), and then treated with 25 μM MG132 or an equivalent volume of the solvent for another 6 h. Whole cell extracts were then produced using RIPA buffer as described in the Materials and Methods section. An aliquot of 30 μg total protein from each extract was loaded onto a 10% polyacrylamide Tris-glycine SDS gel, transferred to nitrocellulose and then probed for p53, Mdm2, MdmX, and β-Actin as described in the Materials and Methods section. (A) Short exposure of the blots to measure the levels of each protein. (B) Longer exposure of the blot for p53 allowed observation of laddering pattern potentially due to ubiquitin modification of this protein in some of the treatments. Treatment of cells with MG132 increases the levels of the p53, Mdm2, and MdmX proteins indicating that each are subject to proteasome degradation although the degradation of p53 in untreated cells appears to be more than in the other treatments consistent with significant levels of laddering of p53 isoforms in the MG132 treated samples.
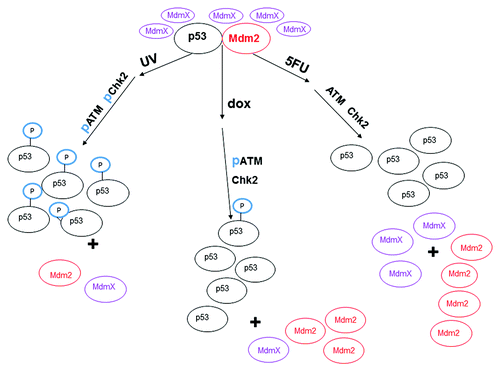
Figure 8. Model of changes in ATM and Chk2 and the effect on the interaction between p53, MdmX, and Mdm2 after treatment of MCF-7 cells with various DNA damaging agents. After treatment of MCF-7 cells with UV, ATM and Chk2 are activated by phosphorylation (represented as “p”) and phosphorylate p53 at serine 15 (represented as “P” sphere on p53). The phosphorylated p53 no longer interacts with Mdm2 which allows accumulation of the p53 protein. ATM is phosphorylated upon treatment of the cells with doxorubicin (dox) for 12 h, but Chk2 is not phosphorylated after this treatment. The p53 protein harvested from dox treated MCF-7 cells is minimally phosphorylated at serine 15 and does not interact with Mdm2. Neither ATM, nor Chk2 are activated in MCF-7 cells treated with 5-fluorouracil (5FU), and the p53 protein harvested from 5FU treated MCF-7 cells is not phosphorylated at serine. The difference in protein levels is approximately represented by the different number of ovals.