Figures & data
Figure 1. Knockdown of USP9X in BxPC3 cells using a Dox inducible lentiviral system. (A) Schematic of the vectors used to inducibly knockdown USP9X. A constitutively active UBC promoter drives expression of a reverse-tet transactivator (rtTA) and also allows for the selection of transduced cells via puromycin. This vector also contains a tet-responsive element that allows for the inducible expression of an RFP marker and a USP9X shRNA sequence. (B) Western blot analysis of USP9X levels in iKD-USP9X-BxPC3 cells grown in the absence or presence of Dox (1 µg/mL) for 6 d. USP9X levels were normalized against GAPDH loading controls, and relative levels are indicated in parentheses. (C) MTT assay examining the growth of iKD-USP9X-BxPC3 cells over time in the absence or presence of Dox (1 µg/mL). The experiment was repeated three times. Data points were averaged and normalized to the day 2 time point, which was set to one. Error bars represent standard deviation, and significant differences between cultures grown with and without Dox are indicated. (D) Representative photomicrographs of iKD-USP9X-BxPC3 cells grown in the absence and presence of Dox for 6 d. (E) Soft-agar growth of iKD-USP9X-BxPC3 cells grown in the absence or presence of Dox. iKD-USP9X-BxPC3 cells grown in the absence of Dox were placed into soft-agar culture conditions, as described in the Materials and Methods. Dox-induced cells were treated with 1 µg/mL Dox where indicated. A scorer, unaware of sample designation, counted the number of colonies observed in the indicated number of high-powered fields (n). Counts were averaged and graphed. Error bars represent standard deviation. The Student t test was used to analyze statistical significance. Growth under anchorage-dependent and anchorage-independent conditions was repeated and similar results were obtained.
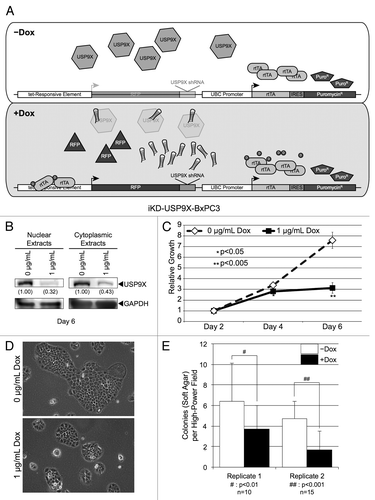
Figure 2. Knockdown of USP9X in Capan1 pancreatic cancer cells. (A) Western blot analysis of USP9X levels in iKD-USP9X-Capan1 cells grown in the absence or presence of Dox (1 µg/mL) for 6 d. USP9X levels were normalized against GAPDH loading controls, and relative levels are indicated in the parentheses. (B) MTT assay examining the growth of iKD-USP9X-Capan1 cells over time in the absence or presence of Dox (1 µg/mL). The experiment was repeated three times. Data points were averaged and normalized to the day 2 time point, which was set to one. Error bars represent standard deviation, and significant differences between cultures grown with or without Dox are indicated. (C) Representative photomicrographs of iKD-USP9X-Capan1 cells grown in the absence or presence of Dox for 6 d. (D) Soft-agar growth of iKD-USP9X-Capan1 cells grown in the absence or presence of Dox. iKD-USP9X-Capan1 cells grown in the absence of Dox were placed into soft-agar culture conditions, as described in the Materials and Methods. Dox-induced cells were treated with 1 µg/mL Dox where indicated. A scorer, unaware of sample designation, counted the number of colonies observed in the indicated number of high-powered fields (n). Counts were averaged and graphed. Error bars represent standard deviation. The Student t test was used to analyze statistical significance. Growth under anchorage-dependent and anchorage-independent conditions was repeated and similar results were obtained.
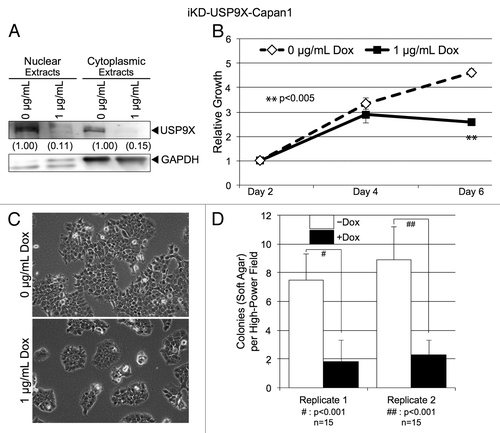
Figure 3. Effects of knocking down USP9X on cell cycle distribution. Cell cycle analysis of iKD-USP9X-BxPC3 cells cultured in the absence or presence of Dox (1 µg/ml) for 4 d. The Telford ReagentCitation46 and FACS analysis (UNMC Cell Analysis Core Facility) were used to determine cell cycle distribution. The Student t test was used to determine P values (n = 2), and error bars represent standard deviations. This experiment was repeated with similar results.
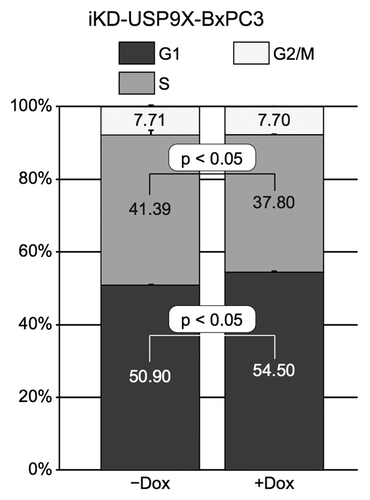
Figure 4. Migration and invasion by iKD-USP9X-BxPC3 cells following knockdown of USP9X. (A) A migration assay was performed as described in the Materials and Methods. iKD-USP9X-BxPC3 cells were grown in the absence or presence (1 µg/mL) of Dox for 48 h and then transferred onto uncoated porous membranes in serum-free medium. Cells were allowed to migrate through the membrane, toward serum-containing medium, for 24 h, before being fixed, stained and counted. The values presented are averages of cells in 14 random, 6.25 mm2 fields. Error bars represent standard error of the mean. The Student t test was used to evaluate statistical significance. This experiment was repeated two additional times with similar results. (B) Similarly, invasion assays were performed as described in the Materials and Methods. Membranes coated with Matrigel were used along with uncoated membranes. Stained cells were counted in 18 random, 6.25 mm2 fields. The bar graph depicts the percentage of invasion (average number of cells per field observed and standard error of the mean for the invasion membranes divided by the average number of cells per field observed on the uncoated membranes). Error bars represent conversion of standard error of the mean. The Student t test was used to test average counts per field for significant difference. This experiment was repeated two additional times, with similar results.
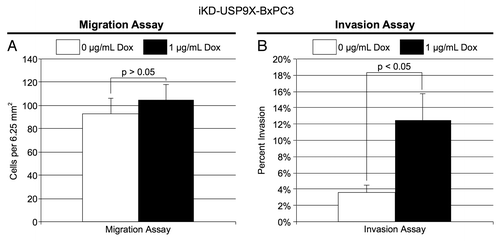
Figure 5. USP9X and ITCH expression in transformed, nestin expressing human pancreatic cell lines. Cell lines were immortalized by expression of the indicated proteins (e.g., hTERT, E6/E7, small T-antigen, KRAS-G12D), as described previously.Citation40 Expression of USP9X and ITCH were examined by western blot analysis. HDAC1 and GAPDH served as loading controls.
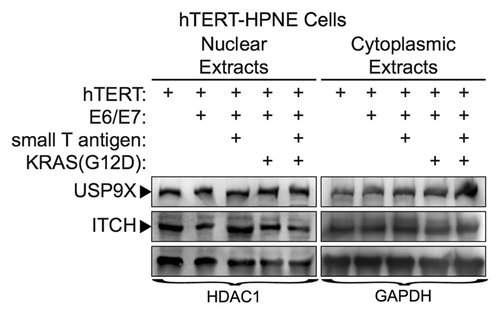
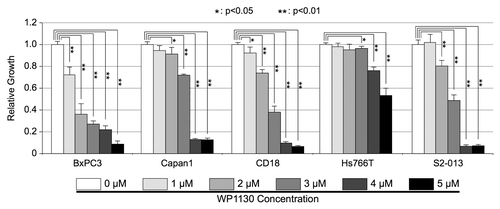
Figure 6. Effects of USP9X inhibitor, WP1130, on in vitro PDAC cell growth. BxPC3, Capan1, CD18, Hs766T, and S2-013, PDAC cells were cultured in the indicated concentrations of WP1130 for 72 h. All conditions tested included the same concentration of the DMSO vehicle. Cell viability was assessed by MTT assay, as described in the Materials and Methods. Measurements from triplicate cultures (n = 3) were averaged and plotted. Error bars represent standard deviations, and the Student t test was used to determine significance. This experiment was repeated and similar results were obtained.