Figures & data
Figure 1. The expression of Ran in pancreatic cancer tissues. (A) Normal pancreatic tissues; (B) Well-differentiated; (C) Poorly-differentiated; (D) Metastatic site in the lymph node; (E and F) Negative controls of normal tissues and pancreatic cancer tissues using PBS instead of primary antibody.
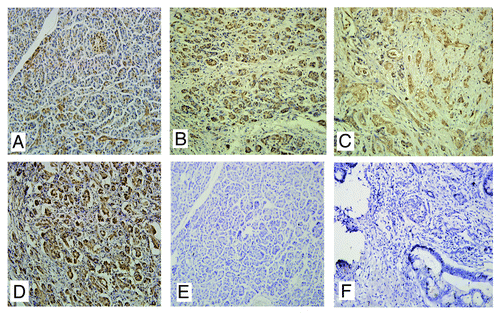
Table 1. Increased Ran expression was statistically correlated with the distant metastasis
Table 2. Expression of Ran in pancreatic cancer and matched lymph node metastases
Figure 2. Ran promoted the migratory and invasive abilities of pancreatic cancer cells in vitro. (A and B) The migratory ability of the cells was evaluated with a wound-healing assay.The wound widths were measured at time 0 and 24 h after wounding, and the closure ratio was calculated in accordance with the following formula: wound closure (%) = (width 0 h) − width 24 h) / width 0 h. These results were then compared with those of the control cells. (C and D) The migratory (8 µm pore polycarbonate filters without matrigel) abilities and invasion (8 µm pore polycarbonate filters with matrigel) abilities of cells were evaluated by transwell assays. Representative image fields of invasive cells on the membrane are shown. The values represent the mean (SEM) from at least three separate experiments, *P < 0.05.
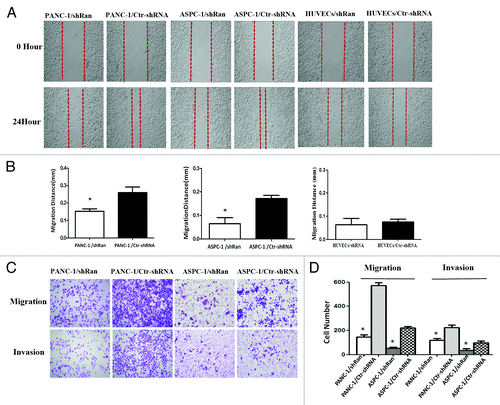
Figure 3. Ran promotes the metastatic ability of pancreatic cancer cells in vivo. Nude mice were tail vein-injected with 1 × 106 PANC-1/ shRan cells and PANC-1/Ctr-shRNA cells. All mice were sacrificed 42 d later after injection of tumor cells and examined for metastatic liver nodules. Representative images of liver tumor metastases. Metastatic loci were identified and marked by arrows; the liver tissues were sectioned serially and then stained with H&E. The number of liver metastasis data are shown in histograms. The values represent the mean (SEM) from at least three separate experiments. *P < 0.05.
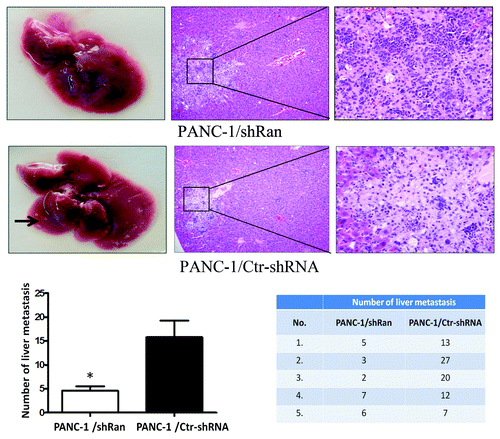
Table 3. mRNA expression of metastasis-related genes after downregulation of Ran
Table 4. mRNA expression of transcription factors after downregulation of Ran
Figure 4. Alteration of gene expression profiling by Ran knockdown in PANC-1 cells. The expression of Ran, AR, and CXCR4 in pancreatic cancer cells was evaluated by western blotting and real-time PCR. β-actin and GAPDH were used as the internal control. *P < 0.05.
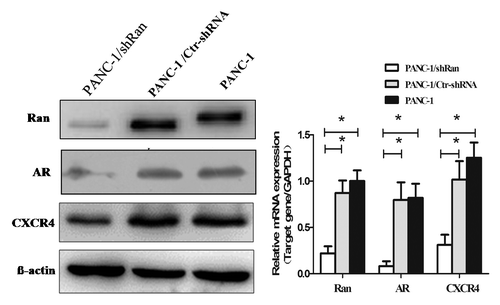
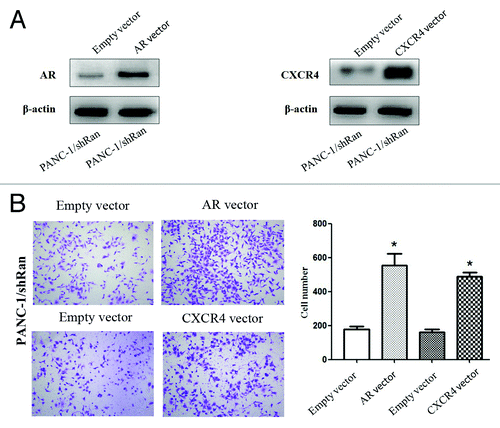
Figure 5. The migratory ability of pancreatic cancer cells after transfection of AR or CXCR4 plasmid into PANC-1/shRan cells. (A) Immunoblotting analysis of AR and CXCR4 expression after transfection of plasmid into cells. β-actin was used as a loading control. (B) Cell migration was increased following transfection of AR and CXCR4 plasmid into PANC-1/shRan cells. *P < 0.05.