Figures & data
Figure 1. The expression levels of Zscan4 in the cancer cell lines analyzed and pulldown assay results between Rap1 and Zscan4. (A) Western results for total cell lysates in both telomerase positive and telomerase negative cancer cell lines using anti-Zscan4 antibody. (B) Zscan4 expression from nuclear and cytoplasmic fractions for SaOS2 and MCF7 cells. β-actin was used as the loading control. (C) The pulldown assay interaction between Rap1 and Zscan4 was analyzed by Coomassie detection (shown in lane 3). The GST negative control showed no signal with purified Zscan4 (lane 4). After GST-Rap1 was immobilized to the beads, eluents from the extensive washing (first and fifth wash) were loaded (shown in lanes 5 and 6). These results demonstrate that unbound GST-Rap1 was clearly removed. Weak signal and no signal were detected in the eluents from the first and fifth washings after purified Zscan4 was incubated with immobilized GST-Rap1 (shown in the lanes 7 and 8). This demonstrated that most of the purified Zscan4 was bound to GST-Rap1 and confirmed a direct interaction exists between Rap1 and Zscan4. GST-Rap1 and purified Zscan4 (shown in lanes 1 and 2) were used as positive controls. (D) Schematic representation of recombinant GST-fused Zscan4 truncations. To identify the minimal domain(s) responsible for the Zscan4-hRap1 interaction, full-length Zscan4 and Zscan4 fragments containing residues (1–126 N-terminal with SCAN BOX-Zscan4, 126–312 middle domain-Zscan4, and 312–433 Zscan4 with Zinc Fingers) were produced as a set of GST–Zscan4 plasmid constructs. (E) The pulldown assays were performed with full-length Zscan4/truncation of Zscan4 and Rap1. Captured proteins were analyzed by western with anti-Rap1 antibody. Signal was detected in the full-length of GST- Zscan4 (Zscan4 1–433) and GST-Zscan4 fragment containing residues 312–433 (Zscan4 312–433). N-terminal of Zscan4 (Zscan4 1–126) and middle domain of Zscan4 (126–312) were negative for signal. This data demonstrated that the C-terminal position of Zscan4 (312–433) was the specific binding site for Rap1.
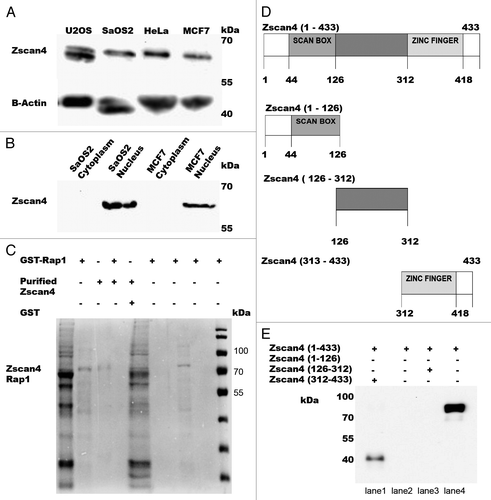
Figure 2. Confocal images of bimolecular fluorescence complementation (BiFC) results in MCF7 cancer cells. DNA was labeled with DAPI (blue, shown in panel 1). Actin filaments were labeled with a fluorescent phalloidin (red, shown in panel 2). Panel 3 is the BiFC signal generated by interaction of the GFP fluorophore components based on proximity. The last panel (panel 4) represents the merged images. (A) GFP signal (green) (shown in the panel 3) was used as a positive control for transfection efficiency in MCF7 cancer cells. The corresponding overlaid images are shown in the last panel. (B) Two vector fragments (pBiFC-VC-155 and pBiFC-VN-173) (shown in panel 3) were used as negative control for BiFC in MCF7 cancer cell line. (C) Two vector fragments (pBiFC-VC-155-TRF1 and pBiFC-VN-173-PinX1) were combined to produce a bimolecular fluorescent complex (green, shown in panel 3). (D) Visualization of Zscan4 and Rap1 complex in MCF7 cancer cells using BiFC analysis (green, shown in panel 3). The images (shown in panel 3) are representative of BiFC analysis for the interaction between Zscan4 and Rap1. Images were generated under an Olympus APON 60X TIRF NA 1.4 objective.
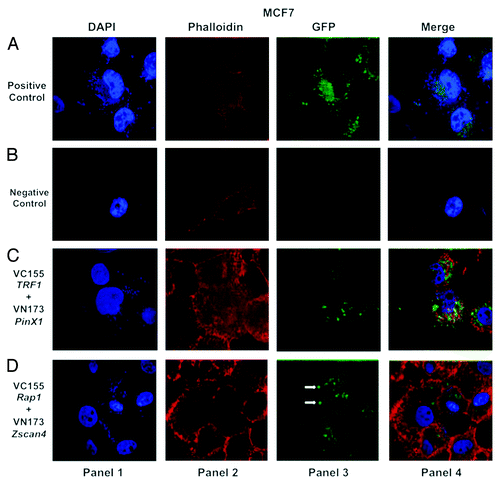
Figure 3. Confocal images of bimolecular fluorescence complementation (BiFC) results in SaOS2 cancer cells. BiFC analysis was performed in SaOS2 cancer cells as described in and reflects the same pattern observed for MCF7 cells. The interaction between Zscan4 and Rap1 (shown in panel 3 of ) occurs in SaOS2 cancer cell line as well. Cells were visualized using the same conditions as described in .
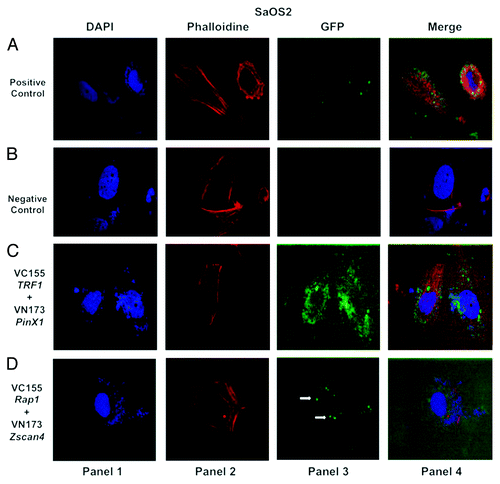
Figure 4. Confocal images of bimolecular fluorescence complementation (BiFC) results in HeLa cancer cells. The BiFC performed in HeLa cancer cells reflects the same patterns observed for MCF7 cells () and SaOS2 cells (). The interaction between Zscan4 and Rap1 (shown in panel 3 of [D]) demonstrates the same relationship in HeLa cancer cells. Cells were visualized using the same conditions as described in .
![Figure 4. Confocal images of bimolecular fluorescence complementation (BiFC) results in HeLa cancer cells. The BiFC performed in HeLa cancer cells reflects the same patterns observed for MCF7 cells (Fig. 2) and SaOS2 cells (Fig. 3). The interaction between Zscan4 and Rap1 (shown in panel 3 of [D]) demonstrates the same relationship in HeLa cancer cells. Cells were visualized using the same conditions as described in Figure 2.](/cms/asset/8fedc4ed-ead0-43b0-b9a9-32740b5bea9d/kcbt_a_10929220_f0004.gif)
Table 1. List of vectors used in this project
Figure 5. Zscan4 expression in cancer cells is dependent on Rap1. Either transfecting with Rap1 siRNA knockdown or overexpressing Rap1 with the addition of Rap1-HA modulated Zscan4 expression in cancer cells. (A) When Rap1 siRNA knockdown was performed, Zscan4 levels decreased. In contrast, Zscan4 levels increased when Rap1-HA (overexpression) was transfected. (B) Transfection efficiency was confirmed by western blot with anti-Rap1antibody. Cancer cells transfected with Rap1 siRNA had no detectable Rap1 signal, but cancer cell expression in cultures transfected with the Rap1-HA had significantly upregulated Zscan4 expression compared with untreated. (C) β-actin was used as the loading control. (D) HA signal was only detected in the overexpressed Rap1-HA lysates.
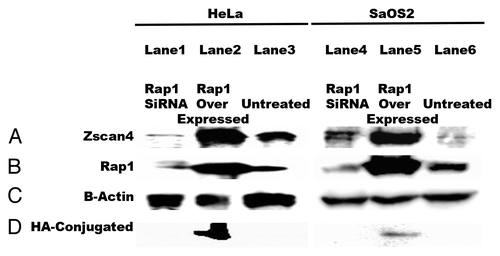
Figure 6. Proposed model of Zscan4–Rap1 interactions in telomere regulation in telomerase-positive cells. (A) TRF2/Rap1 may lose their function of protecting chromosome ends when Zscan4 binds to Rap1. Thus, Zscan4/Rap1 may bind to TRF2 in the shelterin complex of telomeric DNA. (B) The loss of TRF2/Rap1 in the telomere results in an open state (uncapped telomere end) allowing telomerase access to the 3′ overhang.
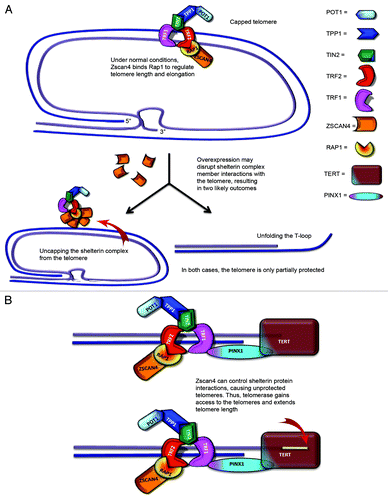
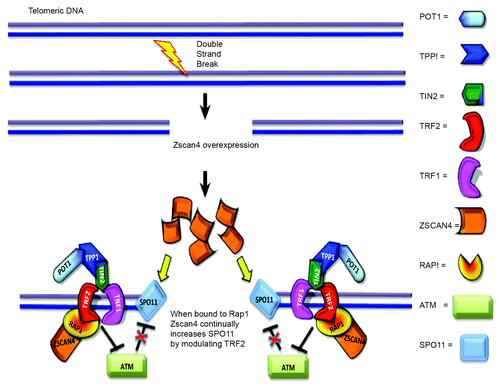
Figure 7. Proposed model of Zscan4–Rap1 interactions in telomere regulation in ALT-pathway cells. When DSBs occur, Zscan4/Rap1 may recruit TRF2 in DNA repair regions of non-telomeric DNA and telomeric DNA. Zscan4 continually increases SPO11 by controlling TRF2 when Zscan4 is bound to Rap1. Alternatively, abundant TRF2, (released from the disrupted shelterin complex through binding of Zscan4/Rap1 to TRF2), might inhibit ATM control of SPO11.