Figures & data
Figure 1. The gatekeeper residue for PERK is methionine 886. (A) Sequence alignment of the PERK ATP-binding pocket with related kinases predicts the conserved gatekeeper residue, shown in red. (B) PP1 inhibitors inhibit the gatekeeper mutant in vitro. Recombinant WT PERK-ΔN and PERK-ΔN M886V (left) or M886A (right) were pre-incubated with 1-NM-PP1 (NM) or 3-MB-PP1 (MB). Kinase was then incubated with recombinant eIF2α in the presence of γ-32P-ATP. Reactions were run on an SDS-PAGE gel and exposed to film. (C) Recombinant WT PERK-ΔN or PERK-ΔN M886G/A/V were pre-incubated with 1-NM-PP1. Kinase was then incubated alone (−) or with p85 substrate in the presence of γ-32P-ATP. Reactions were run on an SDS-PAGE gel and exposed to film. Western blots were performed against total levels to confirm equal loading (Ponceau stains for p85 shown for M886A, M886V).
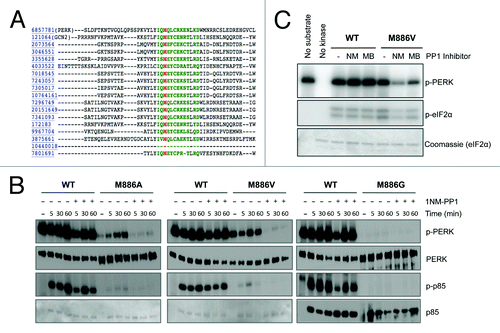
Figure 2. Analog-sensitive PERK alleles are functional in the context of cells. (A) 3-MB-PP1 inhibits PERK M886V. PERK−/− mouse fibroblasts expressing PERK WT or M886V were pre-treated with the indicated doses of 3-MB-PP1, then challenged with thapsigargin (TG) for 1 h. PERK activation was assessed by western blot for PERK and eIF2α phosphorylation. (B) 3-MB-PP1 inhibits PERK M886A. PERK−/− mouse fibroblasts expressing PERK WT or M886A were treated as described in (A). (C) Cells were pre-treated with the indicated doses of 3-MB-PP1, then challenged with TG for 4 h to induce ATF4 and CHOP expression. Cytoplasmic fractions were probed for PERK and p-eIF2α, with total eIF2α to control for equal loading. Nuclear fractions were probed for ATF4 and CHOP, with lamin B as a loading control.
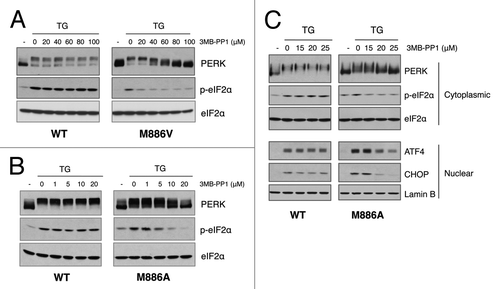
Figure 3. 3-MB-PP1 rescues tunicamycin-sensitivity in wildtype cells, demonstrating an off-target effect. (A) Immortalized PERK−/− mouse fibroblasts stably expressing wild-type PERK were pre-treated with 10 μM (bottom, left) or 15 μM (bottom, right) 3-MB-PP1 for 1 h, or with GSK2606414 as a control. Cells were then acutely stressed with tunicamycin for 30 min and allowed to grow for 6 d. Colonies were stained with Giemsa. (B) Cells of the indicated genotypes were pre-treated with 10 μM 3-MB-PP1 for 1 h, then treated as described in (A).
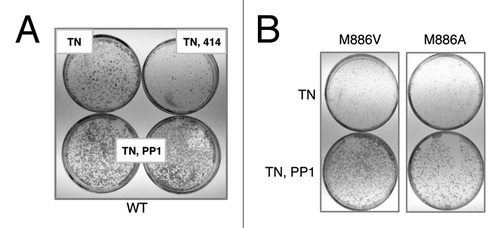
Figure 4. PERK M886A can utilize bulky ATPγS to thiophosphorylate substrate in vitro. (A) The thioP antibody specifically recognizes alkylated, thiophosphorylated PERK substrate. Recombinant WT PERK-ΔN was incubated with eIF2α in the presence or absence of 1mM ATPγS or ATP. Samples were alkylated (PNBM) and assessed by western blot for thiophosphorylated protein using an anti-thiophosphate ester antibody (α-thioP). (B) Only PERK M886A can use bulky ATPγS to thiophosphorylate substrate in vitro. Recombinant WT or M886A PERK-ΔN was incubated with eIF2α in the presence of ATPγS or benzyl (Bn) ATPγS. Reactions were alkylated, then and probed by western blot with the thioP antibody. (C) PERK M886A prefers N6-furfuryl ATPγS. Recombinant WT or M886A PERK-ΔN were used in kinase assays, as described in (B) with the inclusion of the indicated bulky ATPγS analogs.
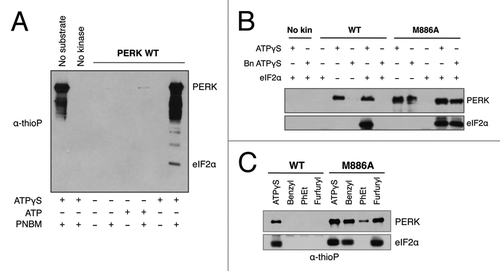
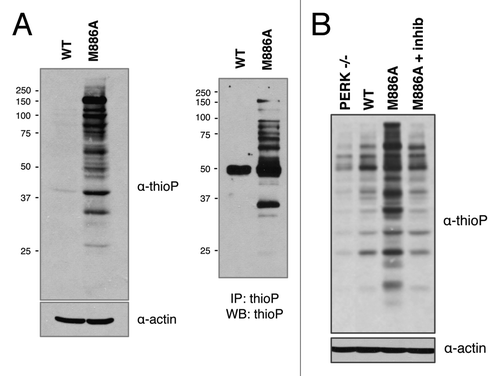
Figure 5. PERK M886A thiophosphorylates substrate in permeabilized cells. (A) PERK−/− cells expressing either PERK WT or M886A were permeabilized, then incubated with thapsigargin and NCitation6-furfuryl ATPγS for substrate labeling. Cells were lysed, and lysates alkylated. Lysates were probed by western blot for total thiophosphorylated substrate (left). Alkylated lysates were then immunoprecipitated with α-thioP (right). (B) Cells expressing M886A were pre-treated with GSK2606414 inhibitor as a control for PERK-dependent activity. The indicated cell types were then treated as described in (A).