Figures & data
Figure 1. Chemical structure of diaporine A (D261, molecular weight = 606). The purity was confirmed to be >98% by HPLC.
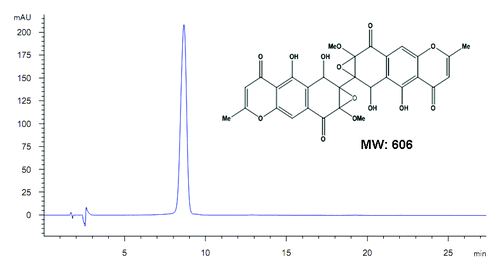
Figure 2. Growth inhibition of human NSCLC cells NCI-H460 and A549 induced by D261. (A and B) Cancer cells were treated with increasing concentrations of D261 for the indicated times. Cell viabilities were evaluated by the CCK-8 assay and denoted as a percentage of vehicle control at the concurrent time point. The bars indicate mean ± SEM (n = 5). (C and D) Cells were stained with CFSE, treated with D261 for 72 h and detected by FACS. Proliferation index was analyzed by modfit software. (E and F) Cells were treated with increasing concentrations of D261 for 24 h and allowed to grow at very low cell density. Colonies were photographed by light microscopy 7 d later and counted as shown. Data are shown as mean ± SEM (n = 3) of one representative experiment. Similar results were obtained in at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared with DMSO control.
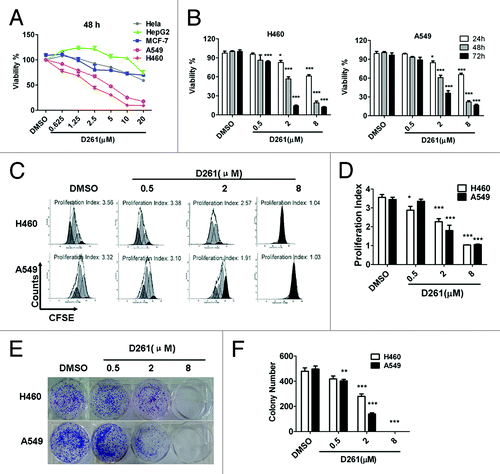
Figure 3. D261 suppresses NSCLC growth in vivo. Female BALB/c nude mice with NCI-H460 xenografts were injected intravenously with D261 or DMSO daily for 3 wk. Body weight of mice (A) and tumor volume (B) were recorded every 3 d. (C and D) Tumor measurement was made after mice were sacrificed. (E) Midkine (MK) expression of the tumor tissue was determined by qRT-PCR. Data are shown as mean ± SEM (n = 7). *P < 0.05, **P < 0.01, compared with DMSO control.
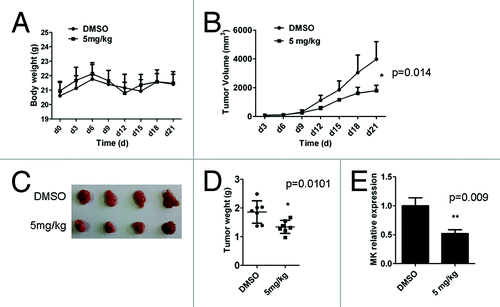
Figure 4. D261 arrests NSCLC cell cycle progression. (A and B) NCI-H460 or A549 cells were treated with various concentrations of D261 for 48 h. Cell cycle was determined by FACS and analyzed by modfit software. (C–E) Effects of D261 on G1/S transition-related molecules were determined by qRT-PCR (C) and western blot (D). Images were representative of three independent experiments. Data are shown as mean ± SEM (n = 3) of one representative experiment. Similar results were obtained in at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
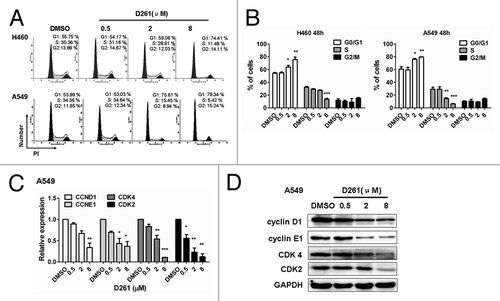
Figure 5. D261 upregulates miR-99a expression in NSCLC cells. NCI-H460 (A) or A549 (B) cells were treated with or without D261 (4 μM) for 48 h, qRT-PCR was performed to determine the relative expression of the indicated micrRNAs. (C and D) MiR-99a fold changes were detected in cells treated with increasing doses of D261 for 48 h. Data are shown as mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 compared with DMSO control.
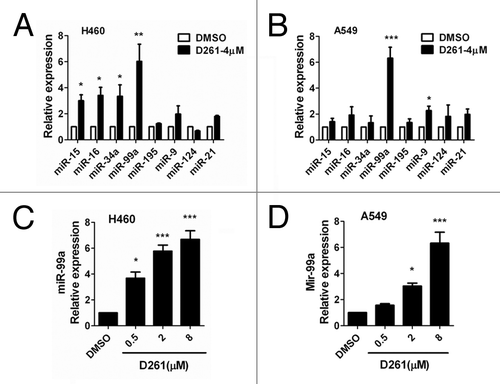
Figure 6. MiR-99a/mTOR is involved in D261-regulated G1/S transition and cell growth in A549. (A) A549 cells were transfected with miR-99a mimics or inhibitors or their negative control oligonucleotides. MiR-99a expression was determined by qRT-PCR analysis. (B) A549 cells were transfected with miR-99a mimics or inhibitors or their negative control oligonucleotides 24 h before treated with or without D261 (4 μM) for another 48 h. Cell viability was assessed using CCK-8. (C) Predicted consequential pairing of target region of mTOR and miR-99a with Targetscan database. (D) Effects of miR-99a and D261 on the activations of mTOR signaling pathways. A549 cells were treated as in (B), protein levels of mTOR and phosphorylation of p70S6K were analyzed by western blot. (E) Expression of cell cycle molecules influenced by miR-99a and D261 were assayed by western blot. Tubulin or GAPDH was performed as a loading control. Images are representative of three independent experiments. Error bars are mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Lip, Lipofectamine RNAiMAX; mi-99a, miR-99a mimics; mi-NC, negative control mimics; in-99a, miR-99a inhibitors; in-NC, negative control inhibitors.
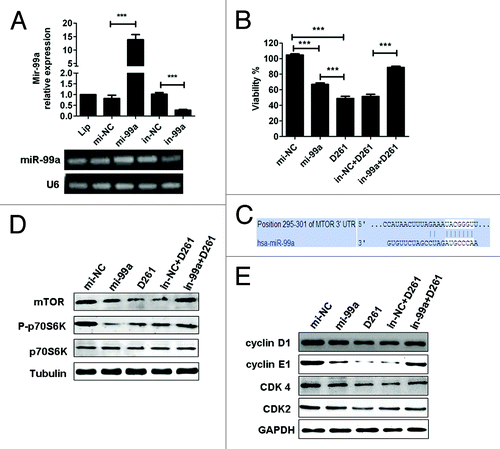
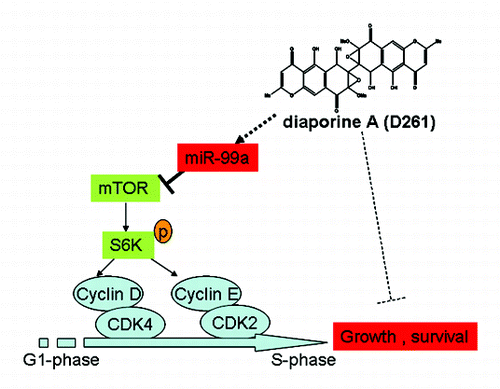
Figure 7. Mechanism of D261 on NSCLC cell growth inhibition. D261 upregulates miR-99a which directly targets mTOR, thus inhibits phosphorylation of S6K (70KD), then induces down-expression of G1/S transition molecules cyclin D/CDK4 and cycline E/CDK2, and results in cell growth inhibition.