Figures & data
Figure 1 Phospho-peptide binding of tandem BRCT domains. (A) Conserved phospho-peptide recognition of BRCA1 BRCT1/2 consists of a phosphate-binding pocket (blue) and +3 binding pocket (brown). The surface representation of BRCA1 BRCT1/2 is shown and the BACH1 peptide pS and +3 side chains are represented as sticks and labeled. (B) pS coordination by BRCA1 (blue) and MDC1 (beige) (left part) and pT coordination by TopBP1 BRCT7/8 (right part). Phosphate contact residues for BRCA1/MDC1 (top/bottom) and TopBP1 are labeled. Equivalent hydrogen bonding and electrostatic interactions are indicated by black dotted lines and additional contacts by TopBP1 R1280 are shown as red dotted lines. (C) +3 phospho-peptide main chain recognition at the BRCT-BRCT interface in BRCA1 (blue), MDC1 (beige) and BARD1 (pink). The equivalent Arg residue in BRCA1/MDC1/BARD1 (top/middle/bottom) is labeled.
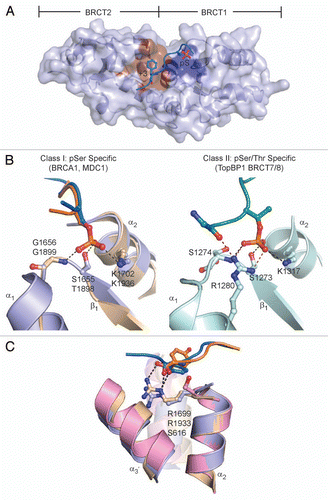
Figure 2 Tandem BRCT domain linker-mediated protein interactions. The inter-BRCT linker is shown in orange and contact regions are highlighted in red. Secondary structure motifs are labeled. (A) Structure of 53BP1 BRCT repeats (gray) in complex with p53 DBD (teal). (B) Structure of the S. pombe Crb2 homodimer. Each protomer is labeled (A and B). (C) LigIV (gray) in complex with XRCC4 homodimer (light cyan).
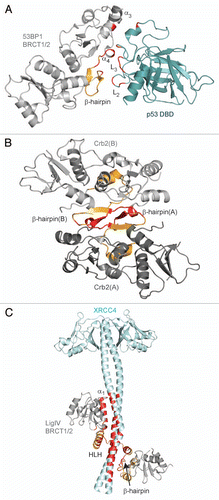
Figure 3 Model of the RFC p140 BRCT domain in complex with dsDNA. (A) Representative ensemble by Kobayashi et al.94 based on the four best structures from HADDOCK. The N-terminal α-helix is shown in purple and the 5′-phosphate is represented as a gray sphere. (B) Surface representation of RFC BRCT domain. Binding surfaces with the DNA major groove (purple) and 5′-phosphate (orange) are marked. (C) Conserved phosphate-binding pocket of RFC BRCT domain, as represented by one model in the ensemble. The 5′-phosphate group and contact residues are represented as sticks and labeled.
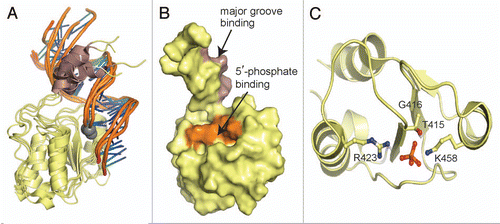
Figure 4 The consensus PAR-binding motif is not conserved in TopBP1 and XRCC1. The PAR-binding motif (cyan) is mapped in the structures of TopBP1 BRCT6 (left) and XRCC1 BRCT1 (right).
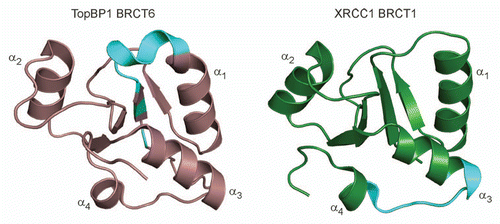
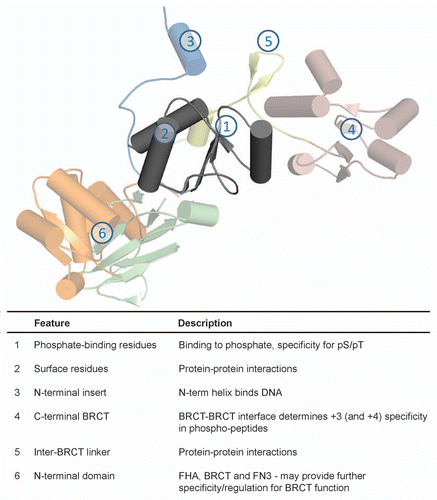
Table 1 BRCT domain architecture found in proteins