Figures & data
Figure 1 NIRF induces G1 arrest. (A) Rapidly growing HEK293 cells were transiently transfected with pEGFP-NIRF (pEGFP-C1-NIRF). After 48 h, the cells were harvested, and the cell cycle profiles were analyzed by FACS as described by Li et al.Citation20 The green fluorescent signal of EGFP was employed as the transfection marker, while PI staining was used to count the DNA contents of the cells. (B) The pEGFP-C1 empty vector was transfected and the cell cycle was analyzed as described for (A).
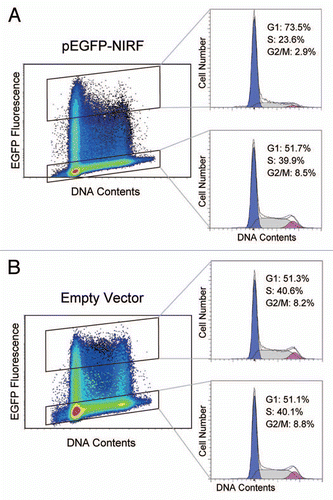
Figure 2 Antibody macroarray screening shows multiple binding partners of NIRF (see Table S1). A lysate of HEK293 cells transfected with Flag-NIRF (p3xFlag-NIRF) was overlaid on the membrane, and the spots for the interactors were visualized by an alkaline phosphatase-conjugated anti-Flag antibody. Enlarged images are also indicated for the areas corresponding to the cell cycle regulators with strong binding signals.
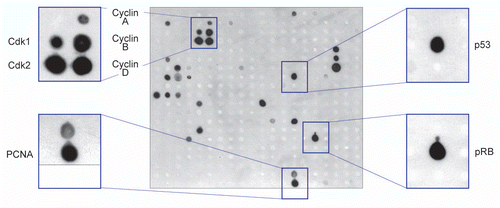
Figure 3 NIRF interacts with cyclins, p53 and pRB. (A) NIRF interacts with cyclins in vivo. COS-7 cells were cotransfected with Flag-NIRF (p3xFlag-NIRF) and myc-cyclins (pCMV-Myc-cyclin A2, B1, D1 or E1) or the empty vectors. MG-132 (5 µM) was added when required. Immunoprecipitation (IP) was performed with an anti-Flag antibody, and analyzed by protein gel blotting (Wesern blot) with the indicated antibodies (left). As a reciprocal IP, myc-NIRF (pCMV-Myc-NIRF) and Flag-cyclins (p3xFlag-cyclins) were used (right). Asterisks represent the ubiquitinated proteins. (B) NIRF interacts with p53 in vivo. HEK293 cells were cotransfected with Flag-NIRF and myc-p53 or the empty vectors. Immunoprecipitation was performed with an anti-Flag antibody, and analyzed by protein gel blotting with the indicated antibodies (left). Reciprocally, myc-NIRF and Flag-p53 were used (right). (C) NIRF interacts with pRB in vivo. HeLa cells were cotransfected with myc-NIRF and Flag-pRB or the empty vectors. Immunoprecipitation was performed with an anti-Flag antibody, and analyzed by protein gel blotting with the indicated antibodies (left). In a reciprocal experiment, Flag-NIRF or empty vector was transfected. Immunoprecipitation was performed with an anti-Flag antibody, and the immunoprecipitates were analyzed by protein gel blotting with the indicated antibodies (right). (D) NIRF interacts with cyclin D1, cyclin E1, p53 and pRB in vitro. The Flag-NIRF protein was expressed in HEK293 cells, and the cell lysate was used for GST pull-down assays with GST, GST-cyclin D1, GST-cyclin E1, GST-p53 or GST-pRB379–928 (large pocket fragment of pRB).Citation81 The SDS-PAGE gel was stained with Coomassie brilliant blue (CBB) while the protein gel blot was probed with an anti-Flag antibody.
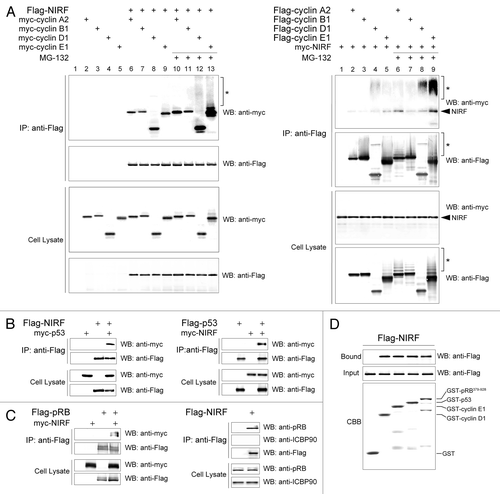
Figure 4 NIRF promotes ubiquitination of cyclins D1 and E1. (A) NIRF ubiquitinates cyclins D1 and E1 in vivo. COS-7 cells were cotransfected with the indicated combinations of plasmids, and then incubated with or without the proteasome inhibitor MG-132. Ubiquitinated cyclins were detected by immunoprecipitation (IP) with an anti-myc antibody, followed by protein gel blot (WB) analysis with the indicated antibodies. Asterisks denote the ubiquitinated proteins, while arrows correspond to nonspecific signals irrelevant to transfection. (B) NIRF ubiquitinates cyclins D1 and E1 in vitro. In vitro ubiquitination assays were performed with the indicated combinations of GST-tagged substrates, E1, E2 (UbcH5a), Flag-ubiquitin (Ub), ATP and either Flag-NIRF or GST-NIRF. After completion of the reaction, the mixtures were subjected to immunoprecipitation with an anti-Flag antibody. The ubiquitinated species (asterisks) were visualized by protein gel blotting with the antibodies against the respective cyclins. Note that the same amount of GST-cyclin substrates was used in each experiment. The arrowheads represent GST-cyclin D1 or E1 proteins captured during the process of immunoprecipitation.
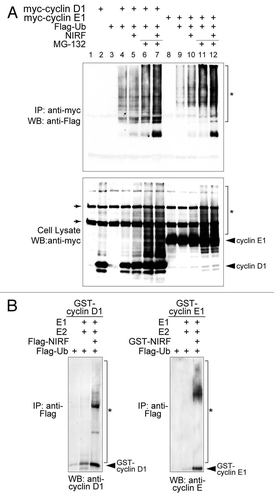
Figure 5 The NIRF interaction network. The PPI information for the core cell cycle proteins (cyclin A2, cyclin B1, cyclin D1, cyclin E1, pRB and p53) were retrieved from the Pathway Commons database.Citation34 Our NIRF interaction data were merged with the retrieved information using the Cytoscape softwareCitation33 as described in the Materials and Methods. The symbols are as follows: yellow, cyclins; yellow-green, CDKs; blue, two major tumor suppressors; green, NIRF-interacting proteins included in the retrieved database information. The thicker black lines indicate interactions demonstrated by more than two methods. The interactions between NIRF and PCNA or HDAC1Citation39 were shown by both the antibody macroarray and the database information. The thinner black lines denote interactions shown by either the antibody macroarray or the pre-existing information in the database. Gray lines indicate other PPI information from the database.
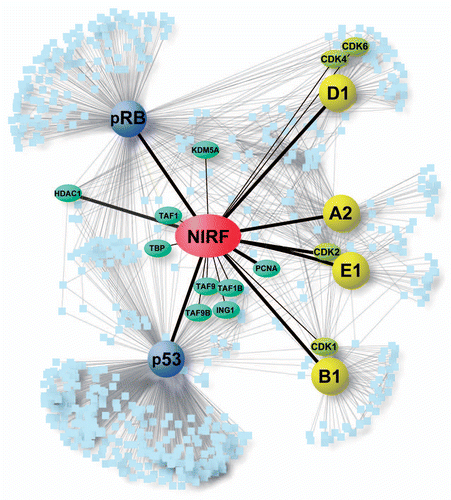
Figure 6 Topological properties of the NIRF interaction network. The NIRF PPI information was subjected to network analyses as described in the Materials and Methods. (A) Shortest path length distribution. The distribution of the shortest path lengths between protein nodes is indicated. The average shortest path length was 2.69. (B) Node degree distribution. The number of protein nodes P(k) with a given degree (k) approximates a power law: P(k) ∼ k−γ; γ = 0.971. Correlation = 0.933, R2 = 0.673. (C) Average clustering coefficient distribution. The average clustering coefficient C(k) of nodes with a given degree (k) follows a power law: C(k) ∼k−γ; γ = 1.210. Correlation = 0.597, R2 = 0.838. (D) Topological coefficient distribution. The topological coefficient TC(k) of nodes with a given degree (k) approximates a power law: TC(k) ∼k−γ; γ = 0.826. Correlation = 0.937, R2 = 0.931. The centrality indices of NIRF were as follows: betweenness centrality, 5.50 × 10−3 (ranked in the top 0.45%, 11th in 2,466 nodes); closeness centrality, 0.475 (top 1.2%, 30th); degree, 19 (top 0.45%, 11th).
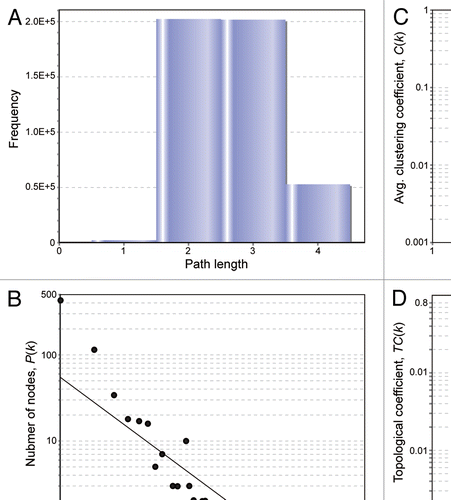
Figure 7 Loss of the NIRF gene in tumor cases. (A) LOH at the NIRF locus in tumors. The genomic DNA was extracted from tumors or the surrounding normal tissues. LOH was determined using the polymorphic markers D9S281 and D9S1852 (left). Summary of the LOH analyses. Fifteen samples were analyzed for each tumor type. LOHs were found in the indicated tumors at varying frequencies. The LOHs at both polymorphic markers coincided in all cases (right). (B) DNA copy number profiles of the NIRF gene in a glioblastoma data set registered in the Oncomine databaseCitation47 (TCGA Brain 2, brain glioblastoma one sample t-test).Citation48 The COPA score at the 10 percentile was −52.222, which was ranked in the top 0.2% of the analyzed genes (37th in 18,823 genes) (p < 1 × 10−8). (C) DNA copy number profiles of the NIRF gene in a lung adenocarcinoma data set in Oncomine (Weir Lung, lung adenocarcinoma one sample t-test).Citation51 The fold change in the mean copy number was −1.084 (p = 1.53 × 10−27).
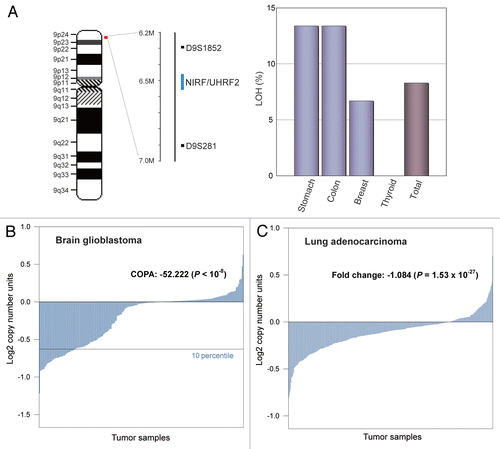
Figure 8 The NIRF gene is the target of a microdeletion in non-small cell lung carcinoma. (A) Chromosomal localization of a commonly deleted region at 9p24. The genes present in 9p21.3–24.3 are indicated according to NCBI MAP Viewer (www.ncbi.nlm.nih.gov/projects/mapview). The green box indicates the NIRF gene, while the gray box indicates the deleted region with a focus on this gene. (B) A recurrent micro-deletion targets the NIRF gene. A non-small cell lung carcinoma data set in Oncomine (Beroukhim Multi-cancer, non-small cell lung carcinoma one sample t-test) Citation69 was analyzed by applying a multigene analysis. The extents of the loss or increase in the DNA copy number are shown in blue and red, respectively, with graded intensities. The gray and green boxes correspond to those in (A).
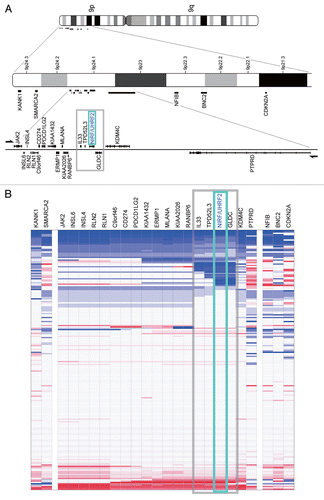
Figure 9 The SNP rs719725 is localized in the vicinity of the NIRF gene. The chromosomal region corresponding to the gray boxes in and B is shown. This region encompasses from IL33 to GLDC, and includes the SNP rs719725, which is localized at 47 kb upstream of the NIRF gene.Citation25
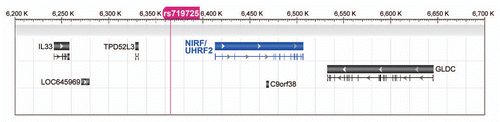
Table 1 Tumors with loss of the NIRF gene showing COPA scores ranked in the top 5%
Table 2 Underexpressed genes in 9p24 in squamous cell lung carcinoma compared with normal tissueCitation70
Table 3 Underexpressed genes in 9p24 in lung adenocarcinoma compared with normal tissueCitation71