Figures & data
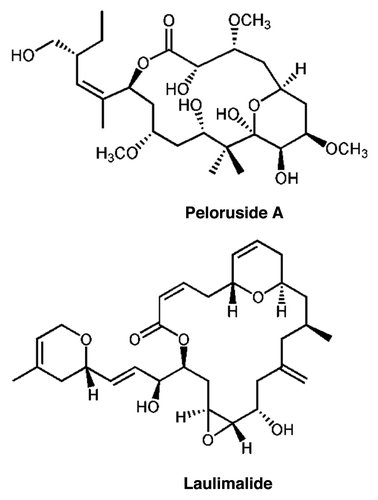
Figure 2 Pel cell lines are resistant to growth inhibition by peloruside but not by paclitaxel or combretastatin A4. Growth and growth inhibition by drugs were determined by the SRB technique as described in methods. Growth is normalized to that observed in the absence of added drug. (A) Relative to the parental cells, Pel A1 and B2 cell lines show greater than 10-fold resistance to growth inhibition by peloruside A. (B) Parental cells and the Pelr lines are equivalently sensitive to growth inhibition by paclitaxel (Taxol). (C) Parental cells and the Pelr lines are equivalently sensitive to growth inhibition by combretastatin A4 (CSA4). Cell lines are identified in the legend in (B). The results shown are from a single experiment. Combined results from multiple experiments are presented in .
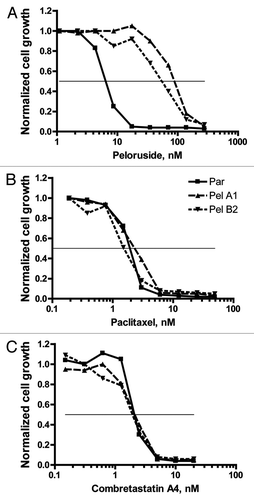
Figure 3 Pelr lines are only resistant to peloruside A/laulimalide binding site drugs. The IC50 for a number of drugs was determined as in for parental and Pelr lines. Fold resistance, calculated as (Pel IC50)/(parental IC50), is presented with error bars representing standard deviation. Resistance was measured for peloruside A (Pel) and five other microtubule stabilizing drugs: laulimalide (Laul), paclitaxel (Ptx), epothilone A (EpoA), dictyostatin (Dictyo) and discodermolide (Disco). In addition four microtubule destabilizing drugs were tested: combretastatin A4 (CSA4), nocodazole (Noc), vinblastine (Vbl) and D24851. Two non-microtubule-targeting drugs were also included: etoposide (VP16), and Romedepsin (or Depsipeptide FK228 = Depsi). Results for all drugs are grouped by Pelr line. The Table below the graph gives the parental cell IC50 and the number or replicates.
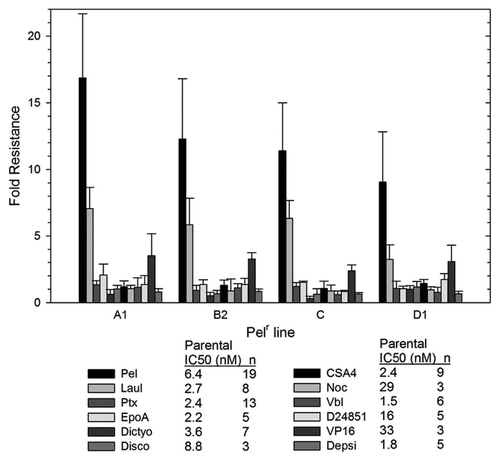
Figure 4 Cell cycle analysis demonstrates that high concentrations of peloruside A induce G2/M accumulation in Pelr lines. Pelr lines and the parental cells were grown in dishes and exposed to varying concentrations of paclitaxel (PTX) or peloruside A (PEL) for 18 h, before trypsinization and staining with propidium iodide to reveal DNA content as described in methods. C-control and drug concentrations were as indicated on the left axis.
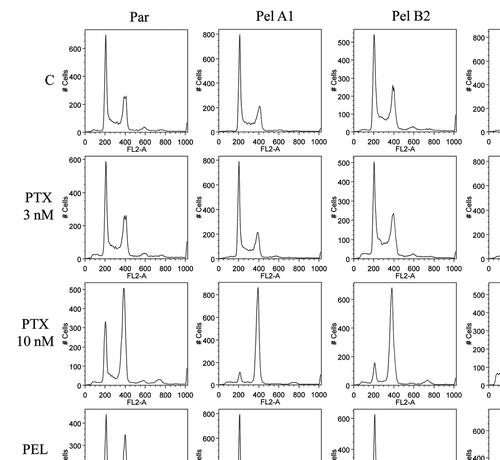
Figure 5 Peloruside A binding site in β-tubulin. (A) Global view of the exterior macrolide binding site on β-tubulin relative to the taxoid binding site on β-tubulin. Protein structure represented as a cartoon model using PBD entry 1JFF, where α-tubulin is colored cyan and β-tubulin pale green. Peloruside A is shown in blue spheres and taxol in red spheres. Exchangeable nucleotide (GDP) is shown as yellow sticks. (B) Closeup view of the binding site showing the position of the Pelr mutations. Binding models showing details of the binding modes for peloruside A (left), based on a reanalysis of docking results and laulimalide (right), adapted from Bennett 2010. Protein structure is rendered in cartoon, highlighting only the binding domain on β-tubulin, and based on PBD file 1JFF as the starting model. Colored secondary structure (orange, yellow, red) represent peptides marking the macrolide binding site by mass shift analysis, from the original determinations of the site (Huzil 2008, Bennett 2010). Ligand structures show only carbon atoms (green) and oxygen atoms (red). Note that both structures orient their long side chains into the cleft defined by in part by A296, R306, N337 and Y340.
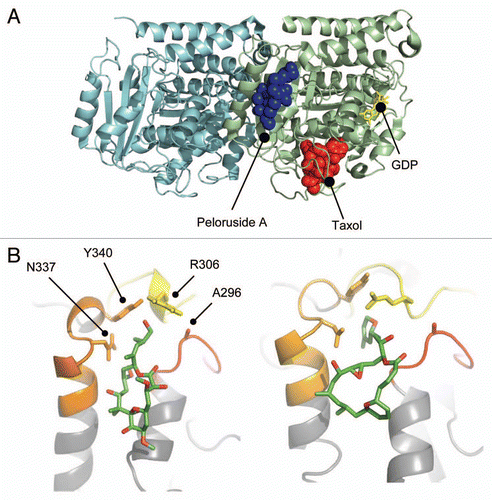
Figure 6 Binding site residues mutated in Pelr mutants are evolutionarily conserved. Residues 296–345 of β-tubulins from a number of sources are aligned to examine sequence conservation. The residues in the binding cavity that are mutated are indicated in bold text and highlighted. (A) Alignment of sequences from human β-tubulin isotypes. (B) Alignment of β sequences from various eukaryotes. Accession numbers: (A) βI NM_178014; βII NP_001060.1; βIII NP_001184110.1: βIVa NP_006078.2; βIVb NP_006079.1; βV NM_032525; βVI NP_110400.1. (B) Human NM_178014; G. gallus AAA49126.1; D. rerio XP_003198209.1; D. melanogaster NP_523795.2; T. thermophila P41352; S. cerevisiae CAA24603; L. tarentolae ABC40567; A. thaliana BAB10059.
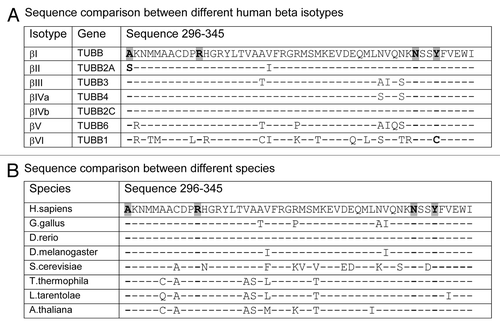
Table 1 Doubling time (hrs) for peloruside A resistant lines
Table 2 Mutations in β-tubulin in Pel lines