Figures & data
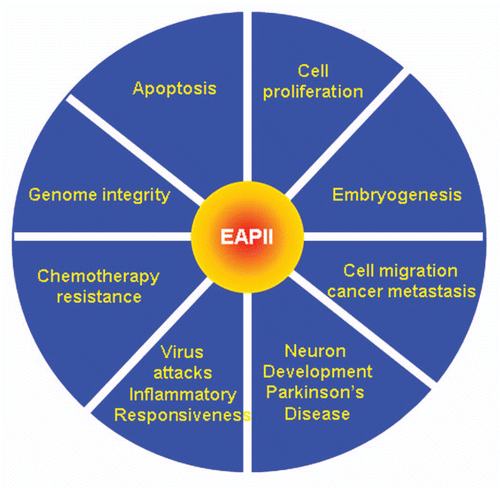
Figure 1 Schematic representation of EAPII protein structure. The UBA domain is indicated in red and the Exo_endo_phos domain in solid blue. Six exo_endo_phos motifs are indicated by the amino acid sequence and the starting and ending number and the functional critical residues are highlighted in red. The potential function of the motifs are: Asn hydrogen bonds of TWN to catalytic Asp of the GDXN motif; Glu of LQE coordinates Mg2+ or Mn2+; GDXN catalytic Asp and Asn hydrogen bonds to scissile phosphate in substrate; and the His of SDH paired with catalytic Asp forms a hydrogen bond to scissile phosphate. The phosphorylation sites (T88 and T92) are also indicated. All amino acid residues are indicated by a one-letter symbol.
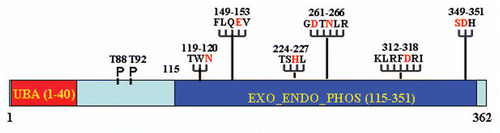
Figure 2 Model of EAPII -mediated transcription modulation: (1) EAPII represses NFκB activity through regulating upstream events of the TRAF-NFκB cascade; (2) EAPII represses transactivation by ETS1 on the MMP1 promoter; and (3) EAPII represses transactivation by Smad3 on Snail1, which is a transcriptional repressor of E-cadherin, subsequently leading to de-repression of E-cadherin transcription. Green lines and arrows denote transactivation and black lines indicate transcriptional repression.
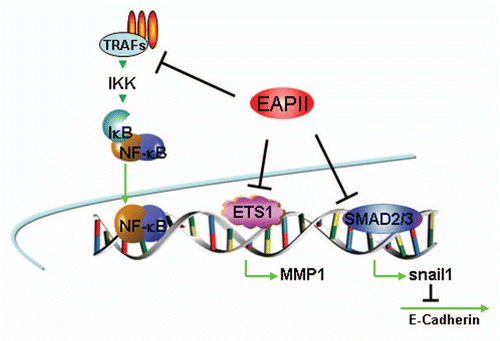
Figure 3 Model of EAPII-mediated signal transduction: (1) EAPII negatively modulates TNFα signaling; (2) EAPII facilitates or represses the JNK-mediated apoptosis pathway, depending on the genotype of DJ-1 protein; (3) EAPII activates MAPK-ERK signaling; and (4) EAPII negatively modulates Nodal signaling through Smad3 association.
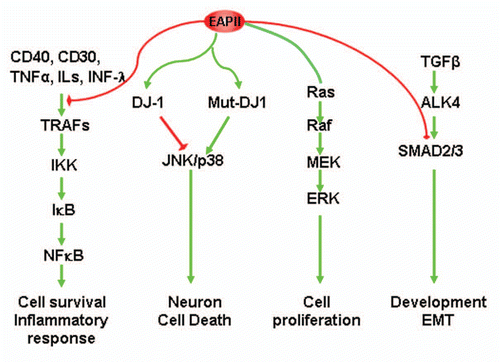
Figure 4 Model of error-free DNA repair function of EAPII: Transit DNA break (cleavable complex) is re-ligated and DNA topological entanglement is resolved (A→B); Stabilizing the transit cleavable complex by TopoII poison results in enzyme-mediated double strand breaks (A→C); TDP2 removes TopoII from protein-DNA adducts and leaves a free phosphate at 5′ of DNA, which is ready for ligation (C→D) and double strand DNA break is repaired (D→B). The solid yellow ovals represent topoisomerase II , and “∼” indicates ester bond. Phosphodiester bond links nucleotide of DNA.
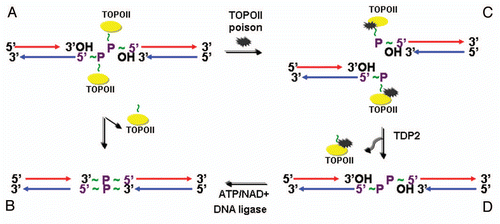
Table 1 EAPII-associated proteins and related biological functions
Table 2 Regulation of EAPII expression