Figures & data
Figure 1 Schematic illustration of the various steps involved in the ubiquitin proteasome pathway and general architecture of the cullin 1 (CRL1) and cullin 4 (CRL4)-based E3 ubiquitin ligases. (A) Ubiquitin molecules (red circles) are covalently attached to the substrate in three consecutive steps. The last step of the reaction is mediated by the specific recognition of the substrate (light magenta), often tagged with a degradation signal, by an E3 ubiquitin ligase (blue) and the transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme (blue) to the substrate. In many cases the degradation signal results in the phosphorylation of the substrate at a specific residues(s). Also shown in blue, is the E1 ubiquitin-activating enzyme involved in charging ubiquitin in the first step of the reaction. The polyubiquitylated protein substrate is directed to the 26S proteasome (light green) where it is degraded proteolytically. (B and C) The scaffold cullin 1 and cullin 4 (cullin 4A or cullin 4B) proteins (light blue) in complex with Rbx1 or Rbx2 small ring finger proteins (purple) form the catalytic core of CRL1 (also known as SCF) and CRL4 respectively. Rbx1/2 proteins recruit the ubiquitin-conjugating enzyme E2 (beige) to mediate the covalent attachment of ubiquitin (red) to the substrate (light green). Skp1 and Ddb1 (damage-specific DNA binding protein 1) (orange) are adaptor proteins for CRL1 and CRL4 respectively and function to bridge the cullin proteins with a number of substrate recognition factors (SRFs) (brown). For Ddb1, one β-propeller domain (BPB) interacts with cullin 4, whereas the two other β-propeller domains (BPA and BPC) make contacts with the SRFs. For CRL1, the SRF are collectively termed F-box proteins because they invariably contain the conserved F-box motif, whereas for CRL4 ubiquitin ligases, SRFs are collectively termed DCAFs (Ddb1 and cullin 4-associated factors). (C) A schematic of the CRL4Cdt2 E3 ligase showing the DCAF Cdt2 (brown) and its ability to recognize substrates only when they are bound to the trimeric PCNA ring (red) encircling DNA (blue helix).
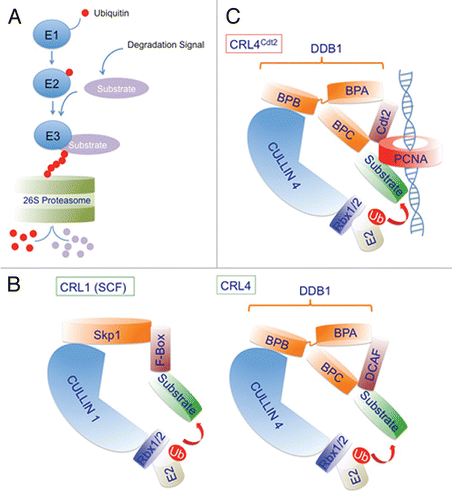
Figure 2 CRL4 ubiquitin ligase regulates several cellular activities via associating with multiple DCAFs. Several DCAFs (Ddb1 and Cul4 associating factors; shown in blue) promote the degradation of various substrates via the CRL4 E3 ubiquitin ligase. The DCAFs, DET1-COP1, Cdt2, CSA and Ddb2 (damage-specific DNA binding protein 2) (shown in blue) recognize the indicated substrates and promote their ubiquitin-dependent proteolysis to regulate various physiological activities such as transcription, chromatin remodeling and DNA repair. The targeted degradation of several of these substrates via CRL4 complexes regulates more than one physiological process.
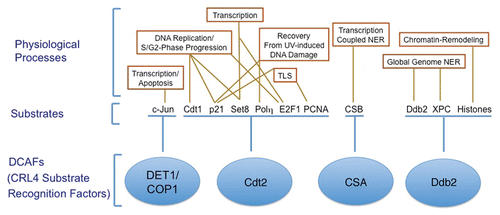
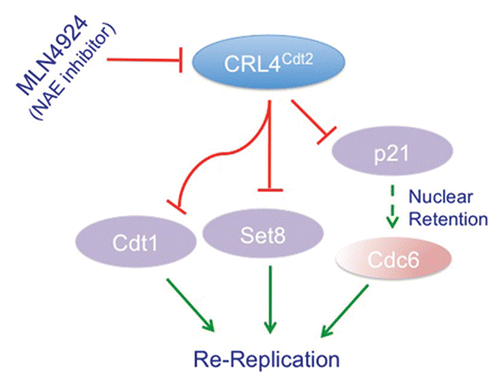
Figure 3 CRL4Cdt2 role in preventing re-replication and genomic instability. A schematic of the various pathways regulated by the CRL4Cdt2 E3 ubiquitin ligase to prevent re-initiation of DNA replication within the same cell cycle (re-replication). By promoting the degradation of the replication-licensing factor Cdt1 in S-phase, CRL4Cdt2 prevents relicensing of replication origins during the S and G2 phases of the cell cycle.Citation31 CRL4Cdt2 also promotes the degradation of the histone H4 lysine 20 methyltransferase Set8 during S-phase thus preventing the premature accumulation of momomethylated histone 4 lysine 20 (H4K20me1) at replication origins, thus inhibiting licensing of these origins.Citation39,Citation41 Additionally, the accumulation of the Cdk inhibitor p21 in cells with inactivated CRL4Cdt2,Citation33,Citation36,Citation37 is hypothesized to inhibit the cytoplasmic export of the licensing factor Cdc6 thereby contributing to re-replication and genomic instability.Citation33 These multiple activities of CRL4Cdt2 are thought to contribute to the re-replication and subsequent apoptosis induced by the novel anti-cancer drug MLN4924, which leads to the inhibition of cullin 4 activities among other cullins.Citation34