Figures & data
Figure 1 CtIP associates with chromatin containing DSBs. (A) Egg extracts containing 35S-Chk1 (lanes 1–4) or 35S-Chk2 (lanes 5–8) and demembranated sperm nuclei were incubated with no checkpoint inducer (lanes 1 and 5) or sperm nuclei treated with 50 µg/ml aphidicolin (lanes 2 and 6), 0.05 U/µl of EcoRI (lanes 3 and 7) or 0.1 U/µl of PflMI (lanes 4 and 8). Nuclear fractions were isolated and analyzed by phosphorimaging and immunoblotting with anti-RPA70 antibodies. Unshifted and shifted forms of Chk1 are indicated with a dash and arrow, respectively. (B) Egg extracts containing sperm nuclei were incubated without checkpoint inducer (lane 1), 50 µg/ml aphidicolin (lane 2) or 0.1 U/µl PflMI (lane 3). Nuclear fractions were subjected to SDS-PAGE and immunoblotted with anti-TopBP1, anti-CtIP, anti-Nbs1, anti-Cdc45 and anti-Mcm2 antibodies. (C) Egg extracts lacking (lane 2) or containing sperm nuclei (lanes 3–5) were incubated without checkpoint inducer (lane 3), 50 µg/ml aphidicolin (lane 4) or 0.1 U/µl PflMI (lane 5). Chromatin fractions were subjected to SDS-PAGE and immunoblotted with anti-TopBP1, anti-CtIP, anti-Nbs1, anti-RPA70 and anti-Mcm2 antibodies. One microliter of extract was loaded in lane 1 as a loading control. (D) Chromatin fractions were isolated from egg extracts lacking (lane 2) or containing sperm nuclei treated as above in (C) (lanes 3–5) or in the presence of 5 mM caffeine (lanes 6–8). One microliter of extract was loaded in lane 1 as a loading control.
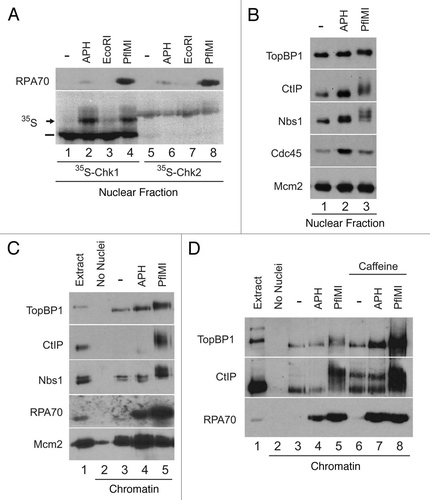
Figure 2 CtIP interacts in a regulated manner with TopBP1 and Nbs1 in Xenopus egg extracts. (A) Control IgG (lane 1) and anti-TopBP1 immunoprecipitates (IP) from interphase extracts incubated in the absence (lane 2) or presence of pA-pT (lane 3) were immunoblotted for CtIP. (B) Control IgG (lane 1) and anti-CtIP IPs from interphase extracts incubated in the absence (lane 2) or presence of pA-pT (lane 3) were immunoblotted for TopBP1. (C) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or CtIP-FLAG (lanes 3 and 4) were incubated in egg extracts in the absence or presence of pA-pT. Beads were retrieved and immunoblotted with anti-TopBP1 antibodies. (D) Control IgG (lanes 1 and 2), anti-CtIP (lanes 3 and 4) and anti-TopBP1 (lanes 5 and 6) IPs from nuclear fractions isolated from interphase extracts supplemented with demembranated sperm nuclei in the absence or presence of 0.1 U/µl of PflMI. Isolated nuclear fractions were subjected to SDS-PAGE and immunoblotted with anti-TopBP1 and anti-CtIP antibodies. (E) Control IgG (lane 1) and anti-Nbs1 IPs from interphase extracts incubated in the absence (lane 2) or presence of pA-pT (lane 3) were immunoblotted for CtIP.
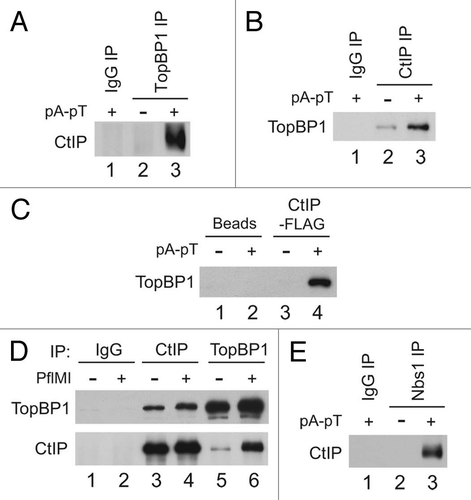
Figure 3 The BRCT I–II region of TopBP1 is necessary and sufficient for association with CtIP. (A) TopBP1 domain architecture. Roman numerals denote BRCT domains of TopBP1. Full-length HF-TopBP1 (WT) and versions of TopBP1 lacking residues 96–282 (ΔI–II) or 360–1485 (BRCT I–II) were produced in Sf9 cells. Location of residues mutated in KKAM mutants is specified in schematic. (B) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or the indicated versions of HF-TopBP1 (lanes 3–8) were incubated in egg extracts in the absence or presence of pA-pT. Beads were retrieved and immunoblotted with anti-CtIP, anti-Nbs1 and anti-FLAG antibodies. (C) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or wild-type (lanes 3 and 4) and KKAM (lanes 5 and 6) versions of HF-TopBP1-BRCT I–II were incubated in egg extracts in the absence or presence of pA-pT. Beads were retrieved and immunoblotted with anti-CtIP, anti-Nbs1 and anti-FLAG antibodies. (D) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or wild-type (lanes 3 and 4) and 2A (lanes 5 and 6) versions of Nbs1-FLAG were incubated in egg extracts in the absence or presence of pA-pT. Beads were retrieved and immunoblotted with anti-CtIP antibodies. (E) Left Part: Representative samples of extracts that were left undepleted (lane 1), mock-depleted with control IgG antibodies (lane 2), depleted with anti-CtIP antibodies (lane 3) or depleted with anti-Nbs1 antibodies (lane 4). Extracts were immunoblotted with anti-CtIP, anti-Nbs1 and anti-RPA70 antibodies. Right Part: Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or wild-type HF-TopBP1 (lanes 3–8) were incubated in the absence or presence of pA-pT in egg extracts that were either mock-depleted with control IgG antibodies (lanes 3 and 4), depleted with anti-CtIP antibodies (lanes 5 and 6) or depleted with anti-Nbs1 antibodies (lanes 7 and 8). Beads were retrieved and immunoblotted with anti-CtIP, anti-Nbs1 and anti-FLAG antibodies. In the CtIP immunoblot, the band in lanes 3, 5 and 7 represents a cross-reacting band, not CtIP signal, as it is present even in CtIP-depleted extracts. (F) Left Part: Representative samples of extracts that were mock-depleted with control IgG antibodies (lane 1) or depleted with anti-CtIP antibodies (lane 2). Extracts were immunoblotted with anti-CtIP antibodies. Right Part: Anti-FLAG antibody beads containing Nbs1-FLAG were incubated in the absence or presence of pA-pT in egg extracts that were either mock-depleted with control IgG antibodies (lanes 1 and 2) or depleted with anti-CtIP antibodies (lanes 3 and 4). Beads were retrieved and immunoblotted with anti-TopBP1 and anti-CtIP antibodies.
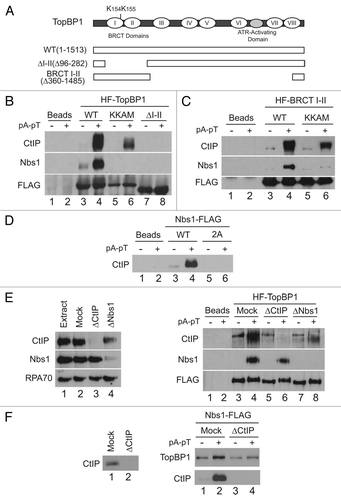
Figure 4 Two distinct regions in the N-terminus of CtIP mediate association with TopBP1. (A) Anti-FLAG antibody beads containing HF-TopBP1 or Nbs1-FLAG were incubated with GST-CtIP (1–349) (lanes 1 and 2) or GST-CtIP(304–856) (lanes 3 and 4) in the absence (lanes 1 and 3) or presence of pA-pT (lanes 2 and 4). Beads were isolated and immunoblotted with anti-GST antibodies. The top part shows in the inputs for the GST fragments. The middle and bottom parts show the TopBP1 and Nbs1 pull-downs, respectively. (B) Anti-FLAG antibody beads containing no recombinant protein (lanes 3, 8, 13 and 18) or wild-type HF-TopBP1 (lanes 1, 2, 4–7, 9–12, 14–17, 19 and 20) were incubated in egg extracts containing full-length 35S-CtIP wild-type (WT) (lanes 1–5), 2AQ (lanes 6–10), Δ25–48 (lanes 11–15) and Δ25–48-2AQ (lanes 16–20) in the absence or presence of pA-pT. The beads were retrieved and bound 35S-labeled proteins were detected by SDS-PAGE and phosphorimaging. (C) Anti-FLAG antibody beads containing no recombinant protein (lanes 3, 8, 13 and 18) or wild-type HF-TopBP1 (lanes 1, 2, 4–7, 9–12, 14–17, 19 and 20) were incubated in egg extracts containing GST-CtIP(1–349) wild-type (WT) (lanes 1–5), 2AQ (lanes 6–10), Δ25–48 (lanes 11–15) and Δ25–48-2AQ (lanes 16–20) in the absence or presence of pA-pT. Beads were isolated and immunoblotted with anti-GST antibodies. (D) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or wild-type HF-TopBP1 (lanes 3–8) were incubated in egg extracts in the absence (lanes 3–5) or presence (lanes 6–8) of tautomycin. Checkpoint activation was induced by the addition of pA-pT (lanes 4, 5, 7 and 8) and PIKK activity was inhibited by supplementing extracts with 5 mM caffeine (lanes 5 and 8). Beads were retrieved and immunoblotted with anti-CtIP, anti-Nbs1 and anti-FLAG antibodies. One microliter of extract was loaded in lane 9 as a loading control. (E) Summary of the capacity of indicated forms of CtIP to interact with TopBP1.
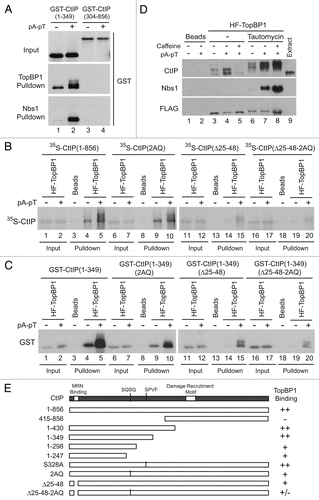
Figure 5 CtIP mediates damage-dependent nuclear accumulation of TopBP1 in response to DSBs in Xenopus egg extracts. (A) Representative samples of extracts that were either left undepleted (lane 1), mock-depleted with control IgG antibodies (lane 2) or depleted with anti-CtIP antibodies (lane 3). Extracts were immunoblotted with anti-ATM, anti-ATR, anti-TopBP1, anti-CtIP, anti-Nbs1 and anti-RPA70 antibodies. (B) Mock-depleted (lanes 1 and 2) and CtIP-depleted egg extracts (lanes 3 and 4) containing 35S-Chk1 were incubated with sperm nuclei in the absence (lanes 1 and 3) or in the presence of PflMI (lanes 2 and 4). Nuclear fractions were isolated and subjected to SDS-PAGE and phosphorimaging to detect radiolabeled Chk1 and also immunoblotted with anti-ATM, anti-ATR, anti-TopBP1, anti-CtIP, anti-Nbs1 and anti-RPA70 antibodies. Unshifted and shifted forms of Chk1 are indicated with a dash and arrow, respectively. (C) Mock-depleted (lanes 1 and 2) and CtIP-depleted (lanes 3 and 4) extracts containing sperm nuclei were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of PflMI. Chromatin fractions were isolated, subjected to SDS-PAGE and immunoblotted with anti-ATM, anti-ATR, anti-TopBP1, anti-phospho[S-1131]TopBP1, anti-CtIP, anti-Nbs1, anti-RPA70 and anti-Orc2 antibodies. ATM is denoted with an arrow, whereas residual ATR signal from previous blotting is marked with an asterisk.
![Figure 5 CtIP mediates damage-dependent nuclear accumulation of TopBP1 in response to DSBs in Xenopus egg extracts. (A) Representative samples of extracts that were either left undepleted (lane 1), mock-depleted with control IgG antibodies (lane 2) or depleted with anti-CtIP antibodies (lane 3). Extracts were immunoblotted with anti-ATM, anti-ATR, anti-TopBP1, anti-CtIP, anti-Nbs1 and anti-RPA70 antibodies. (B) Mock-depleted (lanes 1 and 2) and CtIP-depleted egg extracts (lanes 3 and 4) containing 35S-Chk1 were incubated with sperm nuclei in the absence (lanes 1 and 3) or in the presence of PflMI (lanes 2 and 4). Nuclear fractions were isolated and subjected to SDS-PAGE and phosphorimaging to detect radiolabeled Chk1 and also immunoblotted with anti-ATM, anti-ATR, anti-TopBP1, anti-CtIP, anti-Nbs1 and anti-RPA70 antibodies. Unshifted and shifted forms of Chk1 are indicated with a dash and arrow, respectively. (C) Mock-depleted (lanes 1 and 2) and CtIP-depleted (lanes 3 and 4) extracts containing sperm nuclei were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of PflMI. Chromatin fractions were isolated, subjected to SDS-PAGE and immunoblotted with anti-ATM, anti-ATR, anti-TopBP1, anti-phospho[S-1131]TopBP1, anti-CtIP, anti-Nbs1, anti-RPA70 and anti-Orc2 antibodies. ATM is denoted with an arrow, whereas residual ATR signal from previous blotting is marked with an asterisk.](/cms/asset/c39aee9e-f353-428c-bebe-71b3682b4971/kccy_a_10914711_f0005.gif)