Figures & data
Figure 1 HIF-1α knockdown results in G1 accumulation and p21 induction in U2OS cells. (A) Confirmation of HIF-1α knockdown using U2OS whole cell lysates (WCL) for western blot analysis. Samples were harvested following siRNA protocol and 2 h of DFX treatment. (B) U2OS cells were transfected with nontargeted or HIF-1α siRNA oligonucleotides prior to harvesting for cell cycle analysis using the propidium iodide staining protocol. Student's t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. (C and D) U2OS cells were transfected using nontargeted or HIF-1α siRNA prior to harvest of WCL. Samples were analyzed by western blot using the specific antibodies indicated. (E) U2OS cells were transfected as in (B) and mRNA extracted. HIF-1α, p21, p27 and GLUT3 mRNA were analyzed using quantitative PCR. Graphs depict HIF-1α, p21, p27 and GLUT3 mRNA normalized to actin mRNA and compared to untreated samples. Student t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. Data refers to a minimum of three independent experiments.
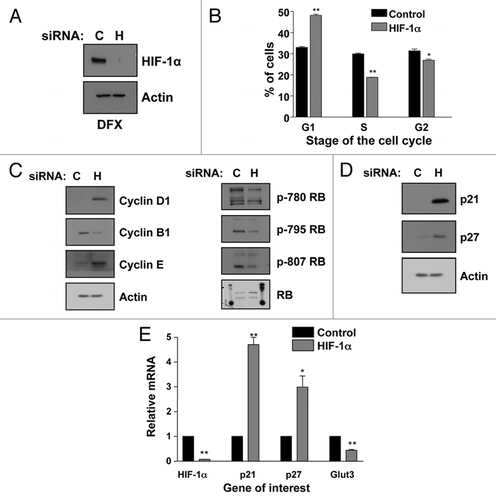
Figure 2 HIF-1α knockdown results in G1 accumulation and p21 induction in HeLa cells. (A) Confirmation of HIF-1α knockdown using HeLa whole cell lysates (WCL) for western blot analysis. Samples were harvested following siRNA protocol and 2 h of DFX treatment. (B) HeLa cells were transfected with nontargeted or HIF-1α siRNA oligonucleotides prior to harvesting for cell cycle analysis using the propidium iodide staining protocol. (C) HeLa cells were transfected using nontargeted or HIF-1α siRNA prior to harvest of WCL. Samples were analyzed by western blot using the specific antibodies indicated. (D) HeLa cells were transfected as in (B) but mRNA was extracted. HIF-1α, p21, p27 and GLUT3 mRNA were analyzed using quantitative PCR. Graph depicts mRNA levels normalized to actin. Student's t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. (E) HeLa cells were transfected with control and HIF-2α siRNA oligonucleotides and mRNA extracted. HIF-1α, HIF-2α, p21, p27 and GLUT3 mRNA were analyzed using quantitative PCR. Graph depicts mRNA levels normalized to actin. Student's t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. Data refers to a minimum of three independent experiments.
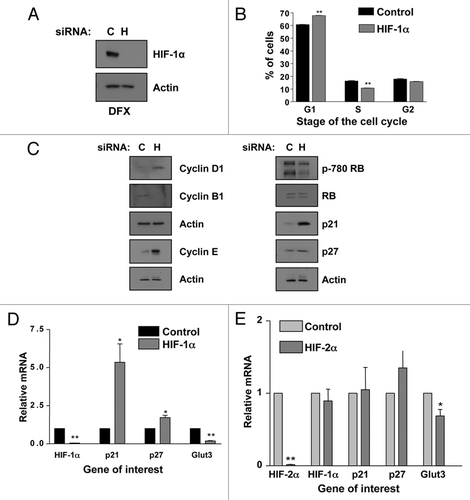
Figure 3 Simultaneous knockdown of HIF-1α and SP1 abrogates p21 induction and partially rescues G1 accumulation. (A) U2OS cells were transfected using nontargeted, HIF-1α, SP1 or both SP1 and HIF-1α siRNA prior to extraction of mRNA. Subsequently, p21, HIF-1α and SP1 mRNA were analyzed using quantitative PCR. Graph depicts mRNA levels normalized to actin. Student t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. (B) U2OS cells were transfected as in A prior to lysis. WCL were analyzed by western blot using the indicated antibodies. (C) U2OS were transfected with control and HIF-1α siRNA oligonucleotides prior to fixation and lysis. ChIP were performed using SP1 and control IgG antibodies. p21 promoter regions were amplified using specific primers and levels of SP1 recruitment were analyzed by qPCR. (D) U2OS cells were treated and processed as in (C), but levels of RNA Polymerase II present at the p21 promoter were analyzed. (E) U2OS cells were transfected as in (A) prior to harvesting for cell cycle analysis using the propidium iodide staining protocol. Student's t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. Data refers to a minimum of three independent experiments.
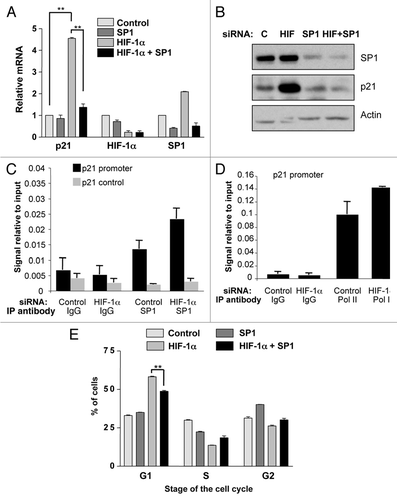
Figure 4 p21 induction and G1 accumulation also occurs in HIF-1α stable knockdowns and non-transformed cells. (A) Two sequences (1 and 2) of shRNA were used to generate U2OS-HIF-1α stable knockdown cells as reported previously in reference Citation61. Confirmation of HIF-1α knockdown in HIF-1α stable cells was performed using western blot analysis. WCL were harvested following 2 h of DFX treatment from non-targeted and HIF-1α stable cell lines. (B) WCLs from U2OS-HIF-1α stable knockdown cell lines were analyzed by western blot using the specific antibodies indicated. (C) U2OS-HIF-1α stable knockdown were harvested for cell cycle analysis using propidium iodide staining protocol. Student's t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. (D) MCF10A cells and (E) HFF cells were transfected with non-targeted or HIF-1α siRNA oligonucleotides prior to mRNA extraction. HIF-1α, p21, p27, SP1 and GLUT3 mRNA were analyzed using quantitative PCR. Graphs depict mRNA levels normalized to actin. Student's t-tests (two tailed) were performed and p-values calculated. *p ≤ 0.05 and **p ≤ 0.01. Data refers to a minimum of three independent experiments.
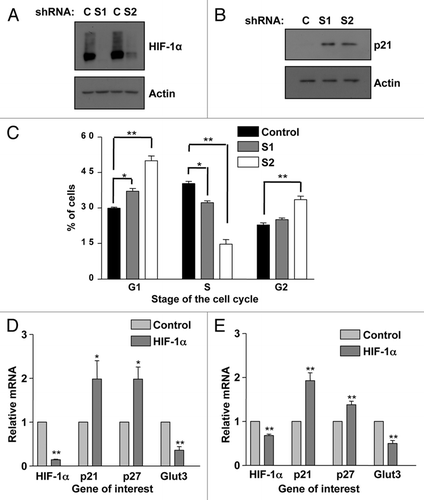
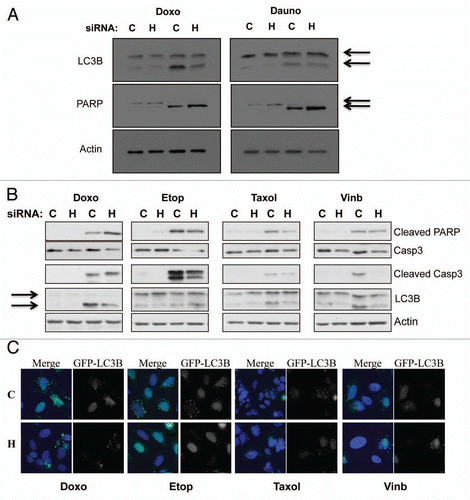
Figure 5 HIF-1α knockdown protects against genotoxic drug induced autophagy, while increasing apoptosis-mediated cell death. (A) U2OS cells were transfected using non-targeted or HIF-1α siRNA prior to 24 h treatment with the genotoxic drugs doxorubicin or daunorubicin as described in the materials and methods. Following treatment WCL were harvested and samples were analyzed by western blot using the specific antibodies indicated. (B) U2OS cells were transfected using non-targeted or HIF-1α siRNA prior to 24 h treatment with the chemotherapeutic drugs doxorubicin, etoposide, taxol and vinblastine as described in the materials and methods. Following treatment WCL were harvested and samples were analyzed by western blot using the specific antibodies indicated. (C) U2OS-GFP-LC3B cells grown on coverslips were transfected using non-targeted or HIF-1α siRNA prior to 24 h treatment with the chemotherapeutic drugs doxorubicin, etoposide, taxol and vinblastine as described in the materials and methods. Cells were fixed, stained with DAPI and analyzed by microscopy for GFP-LC3B foci.