Figures & data
Figure 1 Discovery of miR-365 regulating TTF-1. (A) The top 10 miRNAs predicted by TargetScan (release 5.1) to bind to the 3′UTR of TTF-1 RNA. The list is prioritized based on the context score defined by TargetScan. (B) Luciferase reporter-based screen of candidate miRNAs predicted to bind TTF-1 3′UTR. The psiCHECK-2 (Promega) vector-based luciferase reporter plasmid was cotransfected with individual miRNA oligonucleotides. The negative control (NC) RNA oligonucleotide bears no sequence identity with miRNAs in human, mouse and rat (Dharmacon, CN-001000-01). A validated small interfering RNA against TTF-1 3′UTR (siTTF-1) was included as a positive control (Dharmacon, D-019105-17-0005). The data columns labeled by an asterisk are statistically different from the data of Reporter + NC (p < 0.05, t-test). Y-axis shows the Renilla luciferase signal normalized to (Reporter + NC). Data shown were compiled from three host cells (H441, HeLa and H2170). (C) Seed region of miR-365-binding site and the mutant reporter plasmids. Mutant A includes a 4-bp alteration in the seed sequence (altered bases are underlined), whereas the seed sequence is removed in Mutant B. (D) The seed sequence in the TTF-1 3′UTR critical to miR-365 binding. Wild-type, Mutant A or B reporter plasmid was cotransfected into H441 cells to read out the impact of seed-region alterations in the reporter plasmid. siTTF-1 and wt-reporter were included as controls. An asterisk indicates p-value < 0.05 (t-test) in comparison with the data column of wtReporter + NC.
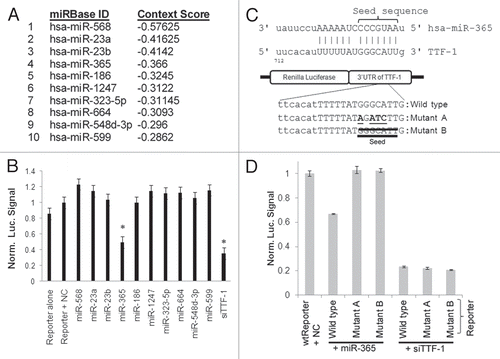
Figure 2 miR-365 suppresses endogenous TTF-1 expression and expression patterns of miR-365 in human tissues. (A) Immunoblotting analysis of H441 cells after transfection of varying amounts of miR-365 RNA oligonucleotide. siTTF-1 RNA oligonucleotide (targeting TTF-1 3′UTR) was included as a positive control. β-tubulin was used as a loading reference. TTF-1 protein levels were quantified and shown below the immunoblot. (B) Immunoblotting analysis of stable retroviral transfectant cells overexpressing mir-365-1, mir-365-2 or mir-365-1m. TTF-1 protein levels were quantified and shown below the immunoblot. (C) RT-QPCR-based RNA expression profiling of miR-365 in 20 normal human tissues. Skel, skeletal muscle; Small, small intestine. (D) Box plot of TTF-1 RNA expression in lung AC samples with highest or lowest miR-365 level. Each category contains 10 samples.
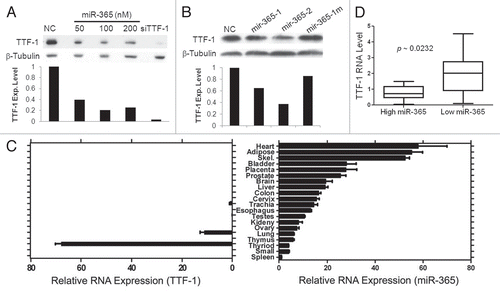
Figure 3 The signaling loop between miR-365 and TGFβ. (A) A time-course study of miR-365 induction by TGFβ. Three lung cancer cell lines and a colon cancer cell strain were subjected to a TGFβ treatment for 3 d. MiR-365 levels were quantified daily as indicated. All data were normalized to the time “0” data point. (B) The RNAs collected from the three lung cell strains treated with TGFβ were analyzed by RT-QPCR for the relative abundance of mir-365-1 and mir-365-2. An asterisk indicates p-value < 0.05 (t-test) in comparison with the respective data column of time 0. (C) TGFβ2 induced by miR-365. Stable transfectant cells (H441) were profiled for TGFβ1∼3 RNA expression by RT-QPCR as indicated. (D) A549 cells carrying empty vector or TTF-1 + 3′UTR were transfected with negative control (NC), miR-365 or siTTF-1. Two days after transfection, the culture media were analyzed using an ELISA assay specific for TGFβ2 (R&D Systems).
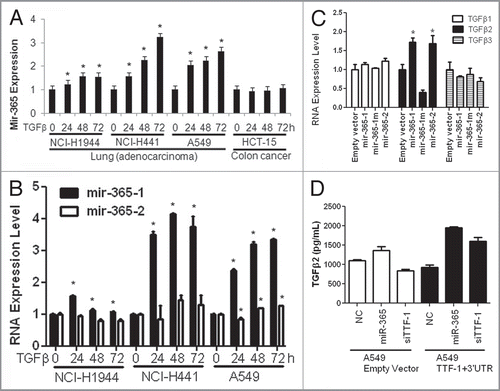
Figure 4 HMGA2 is a target of miR-365. (A) A luciferase reporter assay connected to HMGA2 3′UTR was used in two different host cells to detect the repressive effect by miR-365. Let-7i, a positive control, is known to target HMGA2.Citation37–Citation39 NC, negative control. (B) Endogenous HMGA2 RNA expression was reduced by miR-365. (C) A schematic summarizes the findings of this study. The signaling pathways (a,Citation12 bCitation36 and cCitation27) are known in the literature.
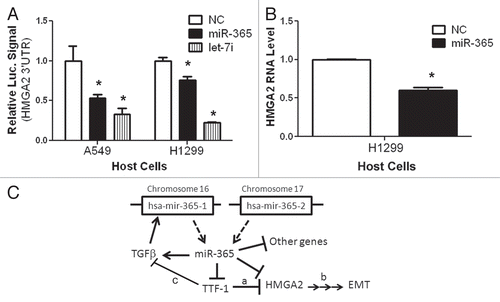
Table 1 RNAs with altered expression in miR-365-expressing H441 cells