Figures & data
Figure 1 P-ΔNp63α bound to the autophagic gene promoters upon cisplatin treatment. ΔNp63α-wt cells were exposed to control medium without cisplatin or medium with10 µg/ml of cisplatin for 12 h. (A) ChIP analysis of the p-ΔNp63α protein binding to the specific region (left parts) and non-specific region (right parts) of the indicated autophagic gene promoter sequences. The amplification of the Input DNA (upper parts) and ChiP-precipitated DNA from untreated cells (middle parts) and cisplatin-treated cells (lower parts) was shown with primers for the specific region and control (non-specific) region. (B) qPCR analysis of the ΔNp63α-enriched binding to the autophagic promoters in ΔNp63α-wt cells and ΔNp63α-S385G cells upon cisplatin exposure. qPCR experiments were performed in triplicate with ±SD as indicated (<0.05). The amount of immunoprecipitated-enriched DNA in each sample (ChIP) is represented as signal relative to the total amount of input chromatin DNA (Input) using the same primers multiplied by 100.
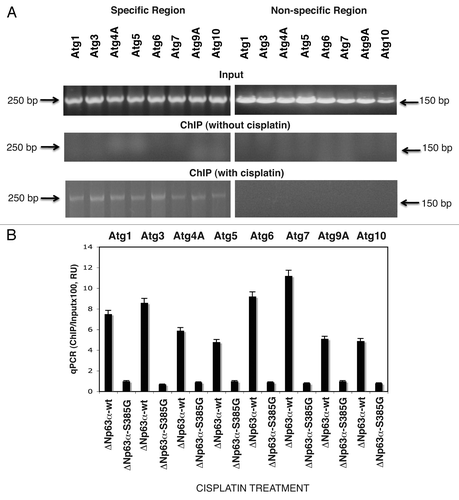
Figure 2 P-ΔNp63α regulated the luciferase activity driven by the autophagic gene promoters upon cisplatin treatment. ΔNp63α-wt cells (A) and ΔNp63α-S385G cells (B) were transfected for 24 h with 100 ng of the LightSwitch_Pro reporter plasmids containing the indicated autophagic promoters or with 100 ng of the control promoter-less reporter plasmid. Cells were exposed to control medium without cisplatin (Con) and medium with 10 µg/ml cisplatin (CIS) for 12 h. RenSP Renilla luciferase reporter activity assays were conducted in triplicate (±SD are indicated, p < 0.05). Data presented as relative to data obtained from the control untreated cells containing the promoter-less reporter plasmid designated as 1.
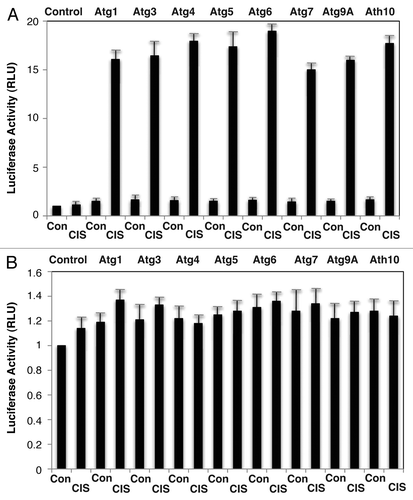
Figure 3 Cisplatin induced specific autophagic proteins. Cells were treated with control medium without cisplatin (Con) or medium with 10 µg/ml cisplatin (CIS) for 16 h. Protein levels were analyzed by immunoblotting with indicated antibodies. The loading levels were tested with an antibody to β-actin. (A and B) ΔNp63α-wt cells. (C and D) ΔNp63α-S385G cells.
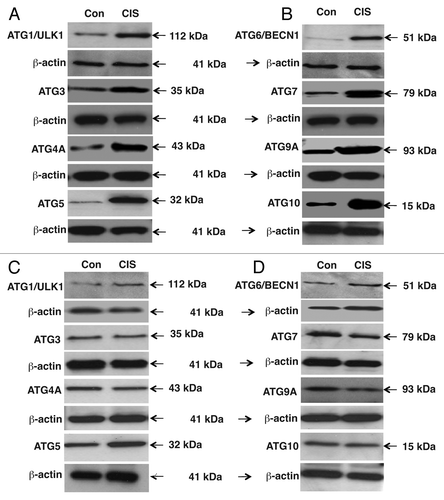
Figure 4 The 3′-UTR sequences for the target autophagic mRNA bound to specific miRNAs upon cisplatin exposure. ΔNp63α-wt cells were transfected with the LightSwitch_3UTR vectors for the specific 3′-UTR sequences as indicated ((A) Atg5/miR-181a and Atg5/miR-374a; (B) Atg6/miR-519a and Atg10/miR-519a; (C) Atg16L/miR-519a and Atg12/miR-630, (D) Uvrag/miR-630 and Uvrag/miR-374a. Cells were also transfected with mock (scrambled) miRNA, and specific miRNA mimics and inhibitors for 36 h. Cells were treated with control medium without cisplatin (Con) or medium with 10 µg/ml cisplatin (CIS) for additional 12 h and then tested for the RenSP Renilla luciferase reporter activity. Measurements (in triplicate) for the luciferase activity (RLU).
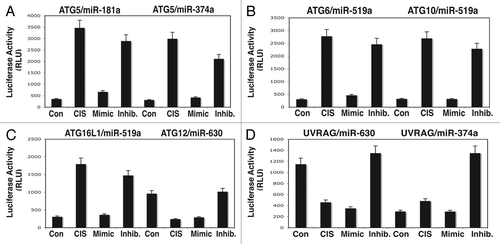
Figure 5 Cisplatin treatment modulates protein levels of autophagic markers through miRNA-dependent regulation. ΔNp63α-wt cells were transfected with the mock (scrambled) miRNA and specific miRNA mimics or inhibitors for 24 h. Cells were treated with control medium without cisplatin (Con) or medium with 10 µg/ml cisplatin (CIS) for additional 16 h and then tested for protein levels of the indicated endogenous autophagic markers. Loading levels were tested using an antibody against β-actin.
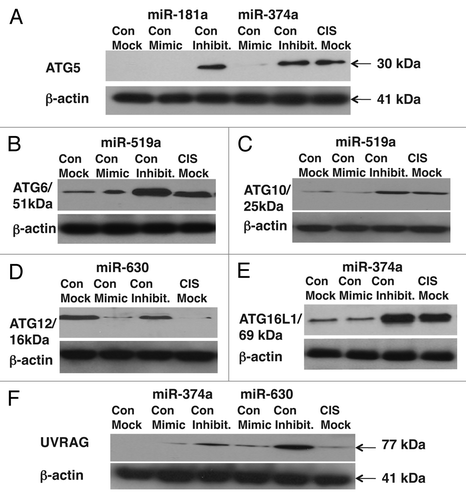
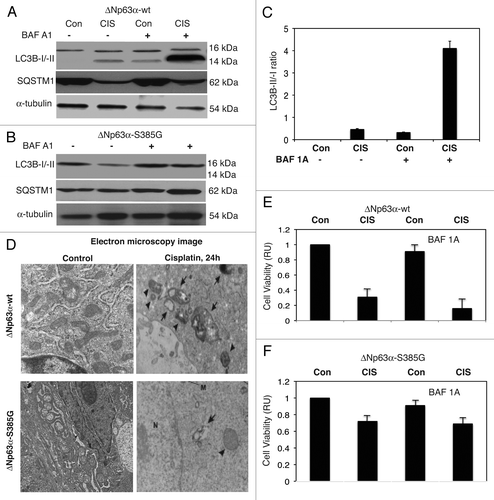
Figure 6 Cisplatin induced autophagic flux in SCC cells. ΔNp63α-wt (A) and ΔNp63α-S385G (B) cells were cultured in the presence (CIS) or absence (Con) of the 10 µg/ml of cisplatin for 16 h and protein levels for LC3B-I, -II, SQSTM1 (p62) and α-tubulin (loading control) were examined using immunoblotting with antibodies to MAP1LC3B (#3868), α-tubulin (#2125) and SQSTM1 (#8025) from Cell Signaling. In some samples, the 100 nM of bafilomycin A1 (BAF A1, #B1793, Sigma) was added for 12 h. (C) Quantitative analysis of LC3B-I/-II conversion. Immunoblots were scanned using Phosphorimager (Molecular Dynamics) and quantified by ImageQuant software version 3.3 (Molecular Dynamics). Values of LC3B-II were expressed as a portion of LC3B-I values defined as 1. The LC3B-II/LC3B-I ratios were plotted as bars using the Microsoft Excel software with standard deviations (±SD) resulting from three independent experiments and three individual measurements of each experiment. (p < 0.05, t-test). (D) ΔNp63α-wt cells (upper parts) and ΔNp63α-S385G cells (lower parts) were cultured in the presence (Cisplatin) or absence (Control) of the 10 µg/ml of cisplatin for 16 h. Cells were trypsinized, fixed and embedded in spur resin. the 90-nm thick sections were cut and examined with a JEOL 1200EX transmission electron microscope (46). Arrows indicate autophagic vacuoles. (E and F) Cell viability assay. ΔNp63α-wt cells (E) and ΔNp63α-S385G cells (F) were cultured in the presence (CIS) or absence (Con) of the 10 µg/ml of cisplatin for 72 h in the presence or absence of BAF A1 (100 nMm 12 h). 104 cells/well in 96-well plates were then incubated in serum-free medium with 5 µg/ml of the 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (American Tissue Culture Collection) in the dark for 4 h at 37°C. Cells were lysed and incubated for 2 h at 37°C, and the measurements (A570 nm to A650 nm) were obtained on a Spectra Max-250 plate reader (Molecular Devices), as described in reference Citation44. Each assay was repeated at three times in triplicate. Diagrams indicated the extent of cell viability expressed as a portion of control represented as 1. The bars are the mean ± SD of triplicate; p < 0.05, t-test.