Figures & data
Figure 1. Effect of anoxia exposure on the relative protein expression of (A) cyclin D1, (B) cyclin D1 localized to the nucleus, and (C) phosphorylated cyclin D1 (Thr286) in liver, kidney and skeletal muscle from aerobic control and anoxic (5 and 20 h) turtles. Representative bands show immunoblot intensity for control and experimental groups. Histograms show normalized expression levels for control vs. 5 and 20 h anoxic conditions; data are means ± SEM (n = 3–4 independent trials on tissue from different animals). * Indicates a significant difference from the corresponding control (p < 0.05).
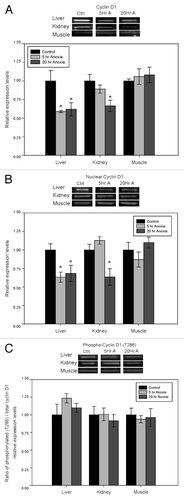
Figure 2. Effect of anoxia exposure on the relative expression of (A) cyclin D1 mRNA, (B) microRNA-16–1 and (C) microRNA-15a in liver, kidney and skeletal muscle from aerobic control and anoxic (5 and 20 h) turtles. Representative bands show RNA transcript levels amplified by RT-PCR. Band intensities from the RT-PCR samples were normalized to either α-tubulin (cyclin D1) or 5S rRNA (microRNA) bands amplified from the same sample. Histograms show normalized expression levels for control vs. 5 and 20 h anoxic conditions; data are means ± SEM (n = 3–4 independent trials on tissue from different animals). * Indicates a significant difference from the corresponding control (p < 0.05).
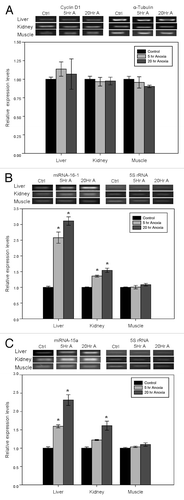
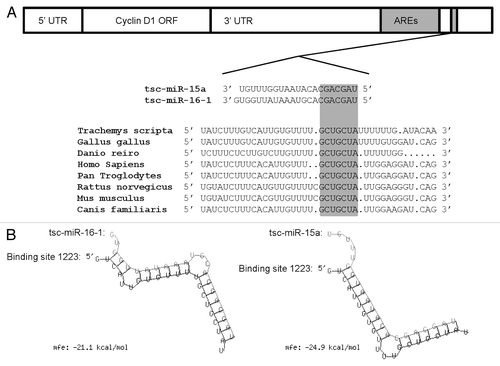
Figure 3. Theoretical binding of microRNA-16–1 and microRNA-15a to a conserved region in the 3′ UTR of the turtle cyclin D1 gene. (A) Conservation analysis of the microRNA-16–1/-15a binding site in the cyclin D1 gene from the red-eared slider turtle (Trachemys scripta elegans), chicken (Gallus gallus), zebra fish (Danio rerio), human (Homo sapiens), common chimpanzee (Pan troglodytes), brown rat (Rattus norvegicus), house mouse (Mus musculus) and domestic dog (Canis familiaris).The seed region sequence (shaded) shows 100% conservation between the eight sequences. (B) Predicted binding structures of both tsc-miR-16–1 and tsc-miR-15a when binding to the 3′UTR of T. s. elegans cyclin D1, as determined from the RNAhybrid program.