Figures & data
Figure 1. Amplification and overexpression of FNDC3B in diverse tumor types. (A) The frequency of amplification is shown for 9 different tumor types along with the overall frequency in > 3,000 diverse tumors using data from Tumorscape (www.broadinstitute.org/tumorscape). High-level amplifications are > 2-fold increase in copy number, focal amplifications are those smaller than one half of the chromosome arm (3q is 104 Mb). (B) Heatmap of copy number alterations along 3q for a large data set of 261 lung tumors.Citation9 The heatmap was created using the Integrated Genomics Viewer (www.broadinstitute.org/igv). (C) Relationship between FNDC3B expression (log2 scale) and DNA copy number in over 400 high-grade serous ovarian cancers in the TCGA data set (http://cbio.mskcc.org/gdac-portal). (D) Detection of FNDC3B protein in amplified and single-copy human liver cells. Immunoblotting with anti-peptide antibodies were used to detect endogenous FNDC3B in HCC cell lines with different copy numbers of FNDC3B as indicated. NexHep is an immortalized non-malignant human hepatocyte.
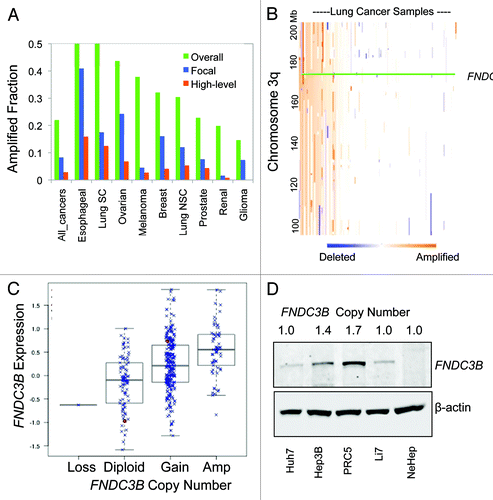
Figure 2. FNDC3B protein localizes to the Golgi network. (A) Fluorescent images of subcellular localization of FNDC3B-GFP fusion protein in C-terminal tail (GFP-C-terminal tail), full length protein (GFP-FNDC3B) and deleted C-terminal lysine (GFP-FNDC3B-ΔK) transfected PHM1 cells. See also Figure S2. (B) Co-staining of FNDC3B (red) and Golgi marker 58K (green) in HCC cell line HepG2. Double positive FNDC3B+ 58K+ cells are shown in a merged view (yellow). (C) Confocal microscopy imaging of endogenous FNDC3B (red) localization in HCC cell line HepG2. (D) AKT, pAKTS473, E-cadherin and Vimentin protein levels in untransfected (U.T), control vector (Vector), FNDC3B overexpressing (FNDC3B) and deleted C-terminal lysine overexpressing (FNDC3B-ΔK) detected by immunoblotting. β-actin was used as a loading control. See also Fig S4.
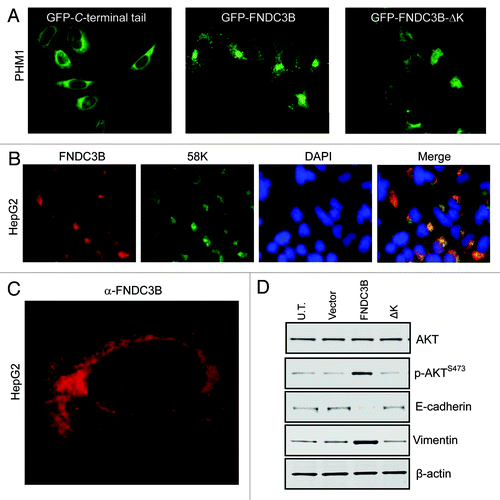
Figure 3.FNDC3B overexpression activates multiple signaling pathways. (A) pRB and PKC-α protein levels in untransfected (U.T.), control vector (E.V.) and FNDC3B overexpressing (FND) PHM1 (left), RK3E (middle) and NMuMG (right) cells were detected by immunoblotting. (B) AKT, pAKTS473 and FNDC3B protein levels in untransfected (U.T.), control vector (E.V.) and FNDC3B overexpressing (FND) NMuMG cells were detected by immunoblotting. β-actin was used as a loading control. (C) AKT, pAKTS473 and FNDC3B protein levels in untransfected (U.T.), control vector (E.V.) and FNDC3B overexpressing (FND) RK3E cells were detected by immunoblotting. β-actin was used as a loading control. (D) AKT and pAKTS473 protein levels EGF treated (treatment with 10ng/ml recombinant EGF for 10 min) in untransfected (U.T.), control vector (E.V.) and FNDC3B overexpressing (FND) PHM1 cells were detected by immunoblotting. (E) Immunofluorescence analysis of Smad4 (red) in control vector (PHM1-Vector) or FNDC3B overexpressing (PHM1-FNDC3B). Smad4/DAPI is shown in a merged view (pink).
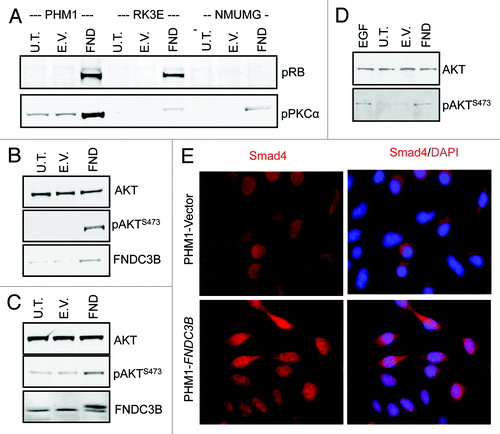
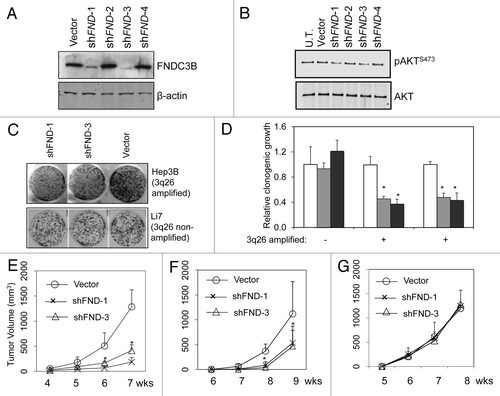
Figure 4.FNDC3B overexpression contributes to tumor maintenance in 3q amplified cell lines. Panel (A) FNDC3B protein levels in PRC5 cell line (3q-amplified) following stable transfection with control vector or shRNAs targeting FNDC3B (shFND1,2,3 and 4) were detected by immunoblotting. shFND1 and shFND3 are effective in silencing FNDC3B protein expression. (B) AKT and pAKTS473 protein levels in PRC5 cell line before (U.T.) or following stable transfection with control vector or shRNAs targeting FNDC3B (shFND1,2,3 and 4) were detected by immunoblotting. (C) Clonogenicity of HepG2 cells (3q-amplified) and Li7 cells (single copy 3q) transfected with shRNAs targeting FNDC3B (shFND-1 and shFND-3) or control vector. (D) Quantification of clonogenicity in three liver cancer cell lines transfected with shRNAs targeting FNDC3B (shFND-1 and shFND-3) or control vector. Results with cells transfected with control vector are shown in white bars, shFND-1 results are shown in light gray bars and shFND-3 results are shown in dark gray bars. Error bars denote ± SD (E) Subcutaneous growth of 5x106 PRC5 cells transfected with control vector or shRNAs targeting FNDC3B (shFND-1 and shFND-3) over a period of seven weeks. n = 10 injections per group. Error bars denote ± SD (F) Subcutaneous growth of 5x106 HepG2 cells transfected with control vector or shRNAs targeting FNDC3B (shFND-1 and shFND-3) over a period of nine weeks. n = 10 injections per group. Error bars denote ± SD (G) Subcutaneous growth of 5x106 Li7 cells transfected with control vector or shRNAs targeting FNDC3B (shFND-1 and shFND-3) over a period of eight weeks. n = 10 injections per group. Error bars denote ± SD.