Figures & data
Table 1. Existing p53-based cyclotherapy studies.
Figure 1. Reversible effects of tenovin-6 and leptomycin B on normal fibroblasts. (A) HNDFs were treated with tenovin-6 or vehicle (DMSO) for 48 h, and then left to recover for 72 h in fresh medium. Graph shows fold increase in cell numbers per field (n = 18) since treatment began. Error bars represent standard deviations. (B) HNDFs were treated with tenovin-6 or vehicle (DMSO) for 72 h, and then left to recover for 96 h in fresh medium before Giemsa staining. (C) HNDFs were treated with various concentrations of LMB or vehicle (absolute ethanol) for 72 h (upper plate) and then grown for 8 d in fresh medium (lower plate) before Giemsa staining.
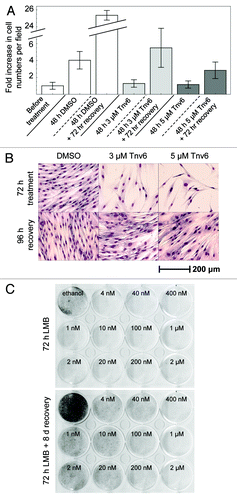
Figure 2. Changes in HNDF cell-cycle distribution in response to small molecules. (A) Cells were treated with the indicated p53 activators for 24 h; and (B) Cells were treated for 24 h and then cultivated for 120 h in fresh medium. DNA synthesis and DNA content were evaluated by measuring BrdU incorporation and propidium iodide (PI) staining by FACS. Values correspond to percentage change compared with untreated controls prior to recovery [i.e., panel A(1)].
![Figure 2. Changes in HNDF cell-cycle distribution in response to small molecules. (A) Cells were treated with the indicated p53 activators for 24 h; and (B) Cells were treated for 24 h and then cultivated for 120 h in fresh medium. DNA synthesis and DNA content were evaluated by measuring BrdU incorporation and propidium iodide (PI) staining by FACS. Values correspond to percentage change compared with untreated controls prior to recovery [i.e., panel A(1)].](/cms/asset/b9449f65-2197-4c09-b43e-d4443a42dbbb/kccy_a_10920254_f0002.gif)
Figure 3. p53 activators protect HNDFs from the adverse effects of S- and M-poisons. Cells were pre-incubated with the indicated small-molecule p53 activators (or vehicle) for 24 h prior to treatment with S- or M-phase poisons for 48 h. After recovery in drug-free medium (4−9 d, depending on drug combination), cells were fixed with methanol−acetone and stained with Giemsa.
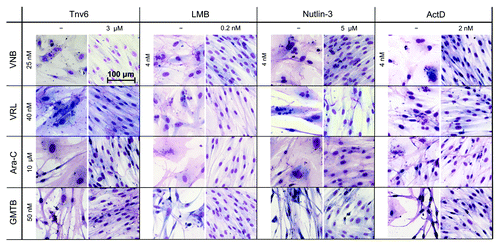
Figure 4. Changes in cell-cycle distribution in response to a cyclotherapy regime combining low dose actinomycin D with gemcitabine. (A) HNDFs and (B) MDA-MB-231 cells were treated as indicated. DNA synthesis and DNA content were evaluated by measuring BrdU incorporation and PI staining by FACS.
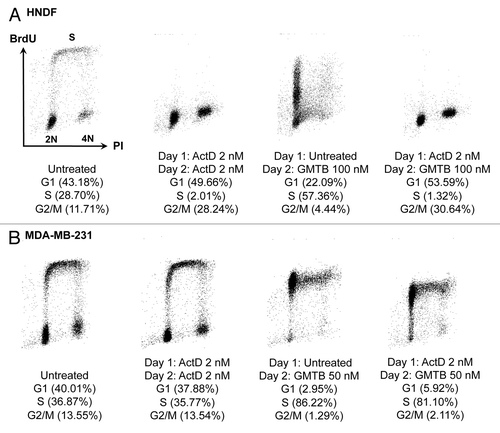
Figure 5. Studies with MDA-MB-231 cells. (A) Cells were subjected to the cyclotherapy protocol described in Materials and Methods, and then stained with Giemsa. (B) Changes in cell-cycle distribution in response to exposure to small-molecule p53 activators for 24 h. DNA synthesis and DNA content were evaluated by measuring BrdU incorporation and PI staining by FACS. Values correspond to percentage change compared with untreated controls [i.e., panel B(1)].
![Figure 5. Studies with MDA-MB-231 cells. (A) Cells were subjected to the cyclotherapy protocol described in Materials and Methods, and then stained with Giemsa. (B) Changes in cell-cycle distribution in response to exposure to small-molecule p53 activators for 24 h. DNA synthesis and DNA content were evaluated by measuring BrdU incorporation and PI staining by FACS. Values correspond to percentage change compared with untreated controls [i.e., panel B(1)].](/cms/asset/d051709f-ebd1-4718-8bbd-c399bb915b14/kccy_a_10920254_f0005.gif)
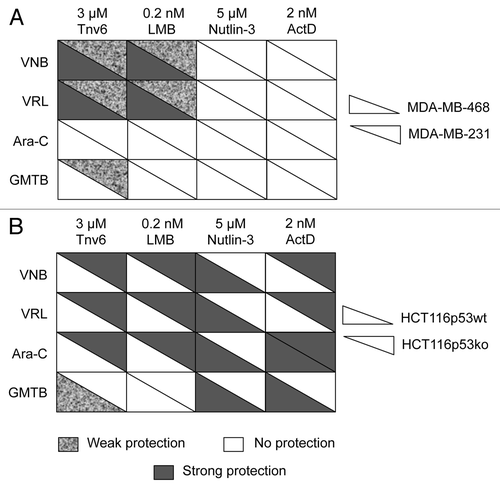
Figure 6. Summary tables for cancer cell lines. (A) MDA-MB-231 and MDA-MB-468 cells and (B) HCT116-p53wt and HCT116-p53ko cells. Cells were subjected to the cyclotherapy protocol described in Materials and Methods. Protection level was decided based upon data recorded after recovery. See also ; Figs. S2 and S5.