Figures & data
Figure 1. Cohesin composition and mechanism of action. (A) Cohesin (blue circle) can entrap DNA segments in trans to mediate sister chromatid cohesion (left) and in cis to form chromatin loops (right). (B) Subunit composition of cohesin and schematic representation of SA1 and SA2 showing their sequence homology.
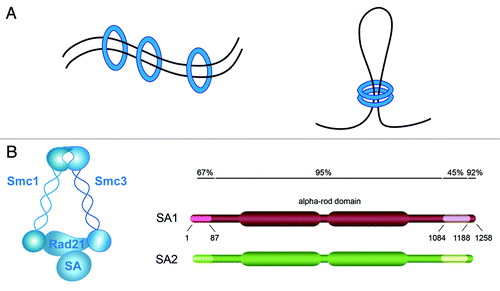
Figure 2. Division of labor between the two cohesin complexes that mediate cohesion along the chromosome. Left: Schematic representation of the distribution of cohesin complexes containing either SA1 and SA2 along the mouse chromosome. Right: Representative images of metaphase chromosomes from MEFs wild type, SA1-null, and wild type treated with SA2 siRNA stained with DAPI (top) or both with DAPI (blue) and a telomeric repeat FISH probe (red). Centromeric cohesion defects are apparent only in the absence of SA2. Aberrant telomere structures (also known as fragile telomeres) reflect defective replication and are visualized as irregularly shaped telomere signals (white arrowhead).
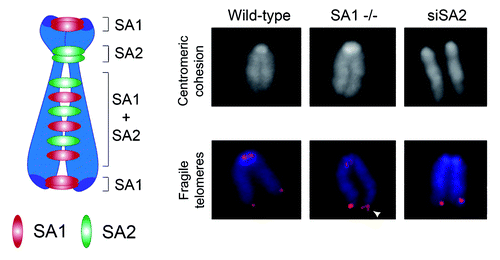
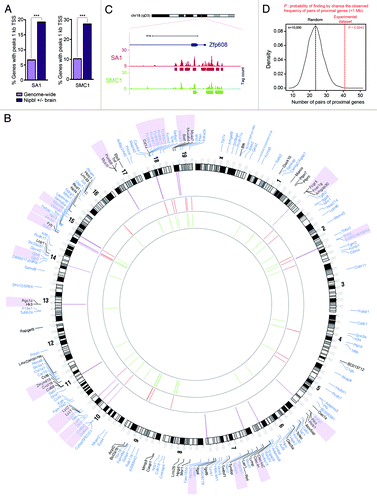
Figure 3. Transcriptional dysregulation in Nipbl +/− and SA1 −/− embryos and CdLS. (A) Percentage of genes with peaks of SA1 or SMC1 located 1kb upstream Transcription Start Site (TSS) for genes genome-wide or genes dysregulated in Nipbl +/− brains. GraphPad Prism 5 software was used to calculate two-tailed χ2 test (with Yates’ correction, ***p < 0.0001). (B) Circos diagram representing chromosomal location of Differentially Expressed Genes (FDR < 0.15) in SA1-null MEFs (black) and genes dysregulated in brains from Nipbl +/− embryos (blue). Regions shorter than 1 Mb containing two or more regulated genes are indicated by purple lines and the genes are shadowed in the same color. Binding sites at gene promoters (up to 1kb upstream the TSS) are depicted for SA1 (red) and SMC1 (green). (C) SA1 and SMC1 peaks at Zfp608 gene, a proposed biomarker of CdLS. This is one of the regions in the mouse genome with highest cohesin peak density. (D) Frequency distribution for pairs of proximal genes (< 1 Mb) in 10,000 random data sets of sample size equivalent to our experimental data set (as defined in B, n = 185). Red line highlights the frequency of pairs of proximal genes observed in the experimental data set.