Figures & data
Figure 1. Mouse cells deficient in clock genes are not sensitive to UV irradiation. Cells were plated at a low density, allowed to attach and then UV irradiated; after colony formation, cells were stained with Giemsa, and percent survival was determined as described in the text. Each data point represents the average of at least three independent experiments and bars signify the standard deviation. (A) Positive arm of the transcription-translational feedback loop (TTFL). Mouse embryonic fibroblasts were derived from mice with the following genotypes: wild-type (squares), Clock−/− (circles) and Bma1l−/− (triangles). (B) Negative arm of the TTFL. Mouse skin fibroblasts were derived from mice with the following genotypes: wild-type (squares), Cry1−/−Cry2−/− (circles) and Per1−/−Per2−/− (triangles).
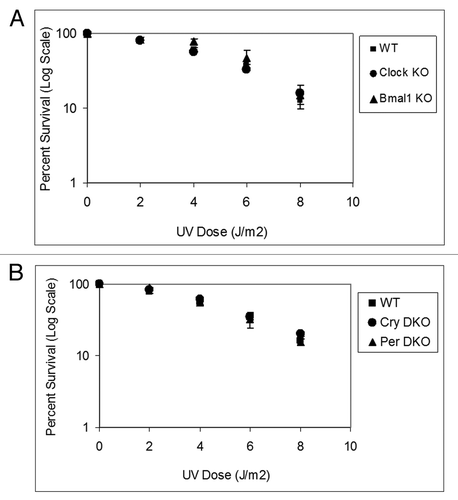
Figure 2. Excision repair capacity of UV-induced photoproducts in mouse cells deficient in positive feedback loop of the core molecular clock. Exponentially growing, primary mouse embryonic fibroblast cells were irradiated with 5 J/m2 UVC, allowed to repair from 0–24 h, lysed and genomic DNA collected for immuno slot blot analysis. (A) An immuno slot blot assay was used to measure (6–4) PP repair at various time points post-irradiation with UV light. The image shows the (6–4) PP signal detected with an anti-(6–4) PP antibody and SYBR gold staining to show equal loading of total genomic DNA. (B) Quantitative analysis of the repair assay from (A) and experiments conducted under identical conditions. (C) An immuno slot blot assay was used to measure CPD repair at various time points, post-irradiation with UV light. The image shows the CPD signal detected with an anti-CPD antibody and SYBR gold staining to show equal loading of total genomic DNA. (D) Quantitative analysis of the repair assay from (C) and experiments conducted under identical conditions. The averages and standard deviations are from at least three independent experiments.
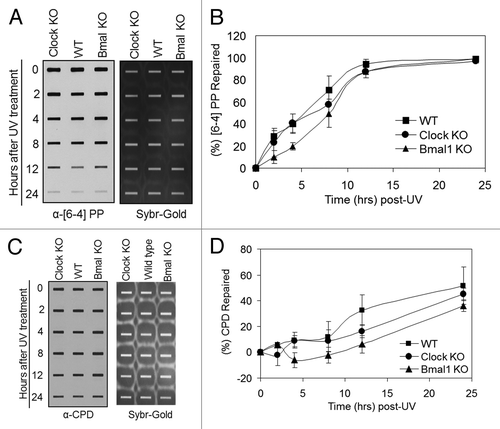
Figure 3. Excision repair capacity of UV-induced photoproducts in mouse cells deficient in negative feedback loop of the core molecular clock. Exponentially growing primary mouse skin fibroblast cells were irradiated with 5 J/m2 UVC, allowed to repair from 0–24 h, lysed and genomic DNA collected for immuno slot blot analysis. (A) Immuno slot blot assay for measuring (6–4) PP repair at various time points post-irradiation with UV light. The image shows the (6–4) PP signal detected with an anti-(6–4) PP antibody and SYBR gold staining to show equal loading of total genomic DNA. (B) Quantitative analysis of the repair assay from (A) and two additional assays conducted under identical conditions. (C) Immuno slot blot assay for measuring CPD repair at various time points, post-irradiation with UV light. The image shows the CPD signal detected with an anti-CPD antibody and SYBR gold staining to show equal loading of total genomic DNA. (D) Quantitative analysis of the repair assay from (C) and two additional assays conducted under identical conditions. The averages and standard deviations are from at least three independent experiments.
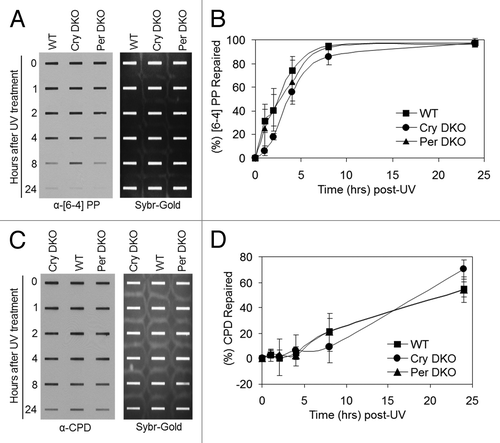
Figure 4. Mouse cells deficient in clock genes are not sensitive to IR irradiation. Cells were plated at low density, allowed to attach and then IR irradiated; after colony formation, cells were stained with Giemsa and percent survival was determined as described in the text. Each data point represents the average of at least four independent experiments and bars signify the standard deviation. (A) Mouse embryonic fibroblasts were derived from mice with the following genotypes: wild-type (squares), Clock−/− (circles) and Bmal1−/− (triangles). (B) Mouse skin fibroblasts were derived from mice with the following genotypes: wild-type (squares), Cry1−/−Cry2−/− (circles) and Per1−/−Per2−/− (triangles).
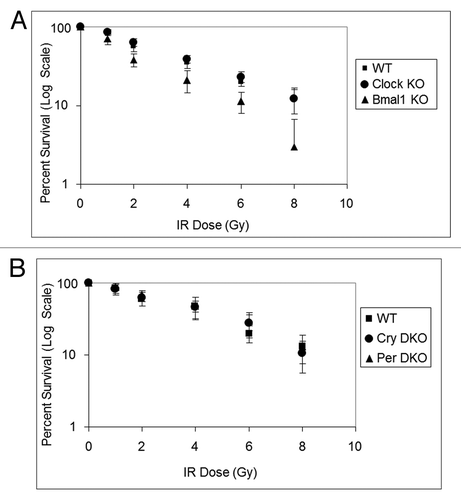
Figure 5. Mouse cells deficient in clock genes are not sensitive to mitomycin C. Cells were plated at a low density, allowed to attach and then treated with mitomycin C (MMC); after colony formation, cells were stained with Giemsa, and percent survival was determined as described in the text. Each data point represents the average of at least three independent experiments and bars signify the standard deviation. (A) Mouse embryonic fibroblasts were derived from mice with the following genotypes: wild-type (squares), Clock−/− (circles) and Bmal1−/− (triangles). (B) Mouse skin fibroblasts were derived from mice with the following genotypes: wild-type (squares), Cry1−/−Cry2−/− (circles) and Per1−/−Per2−/− (triangles).
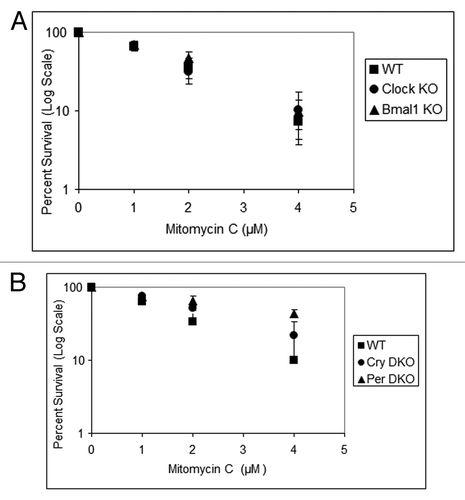
Figure 6. Effect of UV induced Chk1 activation by reduced levels of circadian clock genes in mouse cells. siRNA transfected, primary mouse skin fibroblast cells 72 h later were irradiated with 5 J/m2 UVC, allowed to grow for 0, 0.5 or 1 h and proteins lysates were prepared for immunoblot analysis. (A and C) indicated proteins were analyzed by immunoblotting with appropriate antibodies at various times after UV damage. (B and D) quantitative analysis of the immunoblots. The averages and standard deviations are from at least three independent experiments.
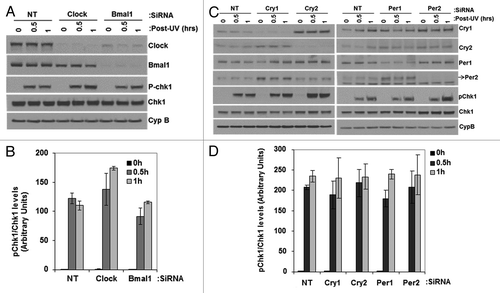
Figure 7. Effect of UV-induced apoptosis by reduced levels of circadian clock genes in mouse cells. siRNA transfected primary mouse skin fibroblast cells were irradiated with 0, 5, 10 or 25 J/m2 UVC, allowed to grow for 24 h and protein lysates were prepared for immunoblot analysis. (A) Protein levels were analyzed by immunoblotting 24 h after UV damage with the indicated antibodies. (B) Quantitative analysis of the immunoblot from three experiments. (C) Protein levels were analyzed by immunoblotting 24 h after UV damage with the indicated antibodies. (D) Quantitative analysis of the immunoblots. Averages and standard deviations are from at least three independent experiments. C-PARP and C-Caspase-3 stand for cleaved PARP and cleaved capsase-3, respectively.
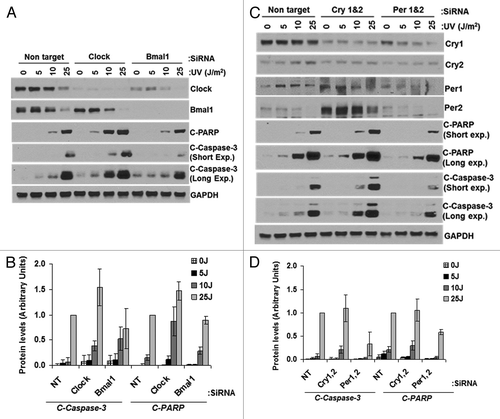
Figure 8. PER1 expression neither sensitizes human lung cancer cells to apoptosis nor affects Chk2 phosphorylation following ionizing radiation (IR). siRNA transfected NCI-H460 human lung cancer cells were irradiated with 0, 10 or 30 Gy of IR and allowed to grow for 48 h, protein lysates were prepared for immunoblot analysis. (A) Protein levels were analyzed by immunoblotting 48 h after irradiation with IR with the indicated antibodies from NCI-H460 cells transfected with either negative control (non-target, NT) or Per1 siRNAs. (B) Quantitative analysis of the immunoblots. (C) Protein levels were analyzed by immunoblotting 48 h after irradiation with IR with the indicated antibodies from NCI-H460 cells transfected with either empty vector (pCDNA3) or a mPer1-overexpressing vector for 72 h. (D) Quantitative analysis of the immunoblots. Averages and standard deviations are from at least three independent experiments. T-PARP and C-PARP stand for total PARP and cleaved PARP, respectively.
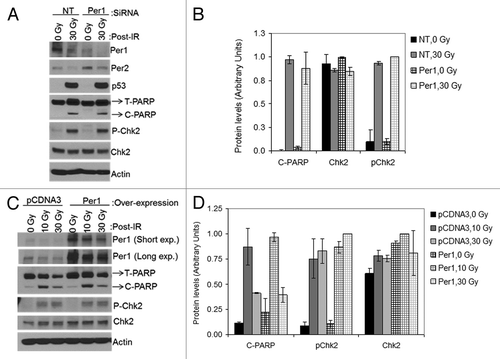
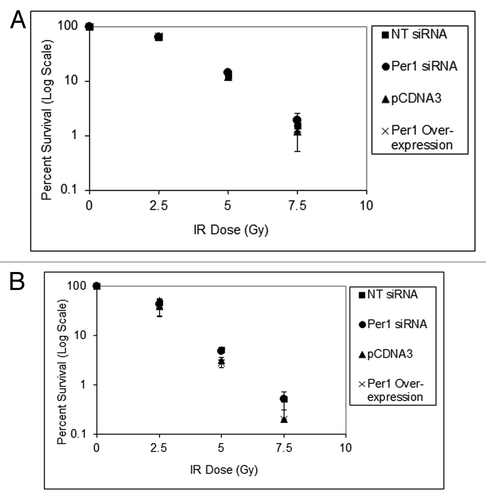
Figure 9. PER1 expression does not sensitize human cancer cells to ionizing radiation in a colony formation assay. Cells were transfected with indicated siRNAs or expression vectors and exposed to IR, and IR survival was measured by colony formation assay. (A) Colony formation assay results for NCI-H460 human lung cancer cells. After 48 h of siRNA treatment, cells were trypsinized and plated at low density (500 cells per dish) in 6-well culture dish plates. After allowing cells to attach for 8 h, they were irradiated with the indicated fluence of IR and allowed to grow in fresh growth media for 7–10 d. Colonies (> 50 cells) were counted using a digital counter after staining with Giemsa and the surviving cell fraction was calculated. (B) Colony-formation assay results for HCT-116 human colon cancer cells. After 48 h of siRNA treatment, cells were trypsinized and plated at low density in 6-well plates. The results are the average of three biological experiments and error bars represent the standard deviation of the mean.