Figures & data
Figure 1. Knockdown of B-Myb reduces the fraction of S-phase cells. (A) HepG2 cells transfected with control siRNA or with two different B-Myb-specific siRNAs. Cells harvested 48 h after transfection were analyzed by western blotting with antibodies against B-Myb and β-actin. (B). HepG2 cells transfected with control or B-myb siRNAs were labeled for 5 min with EdU to visualize S-phase cells. Additionally, cells were stained with antibodies against histone H3 to visualize the total number of nuclei. (C). Quantification of EdU-positive cells. The bars show the percentage of EdU-positive cells in the total cell population, determined by counting 1,000 cells from different photographic fields and several independent knockdown experiments. Thin lines show standard deviations. The asterisks indicate statistical significance (** p < 0.01, Student’s-t-test).
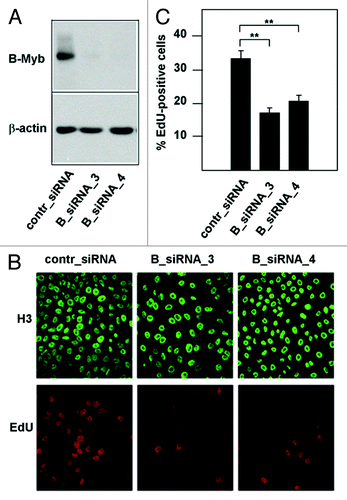
Figure 2. Effect of B-Myb knockdown on S-phase progression. (A). HepG2 cells transfected with B-Myb-specific and control siRNAs were synchronized by mimosine treatment. The cells were then released into S-phase for the indicated times, followed by flow cytometry to examine the progression through S-phase. The white and black arrows mark the G1 and G2 DNA content, respectively. (B). The successful knockdown of B-Myb was confirmed by western blotting.
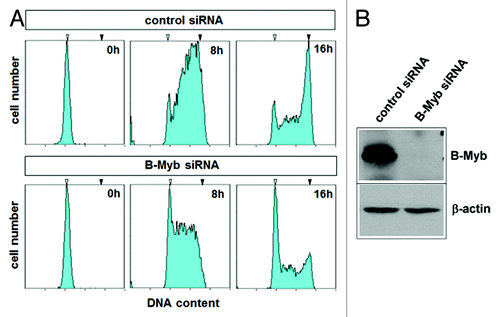
Figure 3. Construction of a DNA-binding defective and siRNA-resistant B-Myb mutant. (A). Schematic illustration of the DNA-binding deficient and RNAi resistant B-Myb mutant. The nucleotide and amino acid sequences of the wt and mutant B-Myb at the B-myb_siRNA4 target region are shown. (B). Equal amounts of GST-B-Myb-wt and GST B-Myb-N174A proteins were subjected to an EMSA experiment using a Myb-binding site oligonucleotide (top). Aliquots of the protein extracts were analyzed by SDS-PAGE and stained with Coomassie brilliant blue to confirm equal protein concentration (bottom). (C). HepG2 cells were first transfected with expression vector for B-Myb (N174A) mutsiRNA_4. Equal aliquots of these cells were subsequently transfected with control siRNA, B-myb_siRNA3 or B-myb_siRNA4 and analyzed by western blotting 24 h later.
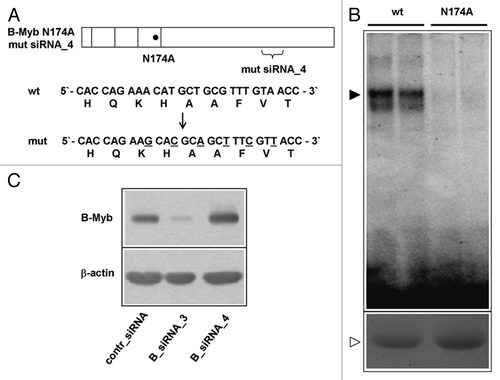
Figure 4. Decreased S-phase entry after B-Myb knockdown is rescued by a DNA-binding deficient and RNAi-resistant B-Myb mutant. (A). HepG2 cells expressing doxycyclin-inducible B-Myb N174A mutsiRNA_4 were grown in the absence or presence of doxycyclin, followed by immunofluorescence analysis. Only the cell nuclei are visible. (B). The cells were additionally transfected with control or B-Myb-specific siRNA_4, followed by western blotting with antibodies against B-Myb and β-actin. (C). HepG2 cells expressing doxycyclin-inducible B-Myb N174A mutsiRNA_4 were grown in the presence or absence of doxycyclin and transfected with the indicated siRNAs. The cells were then analyzed for the percentage of EdU-positive cells (top) and by real-time PCR for the expression of the cdc2 and cyclin B1 mRNAs. The asterisks indicate statistical significance (* p < 0.05, ** p < 0.01, Student’s-t-test).
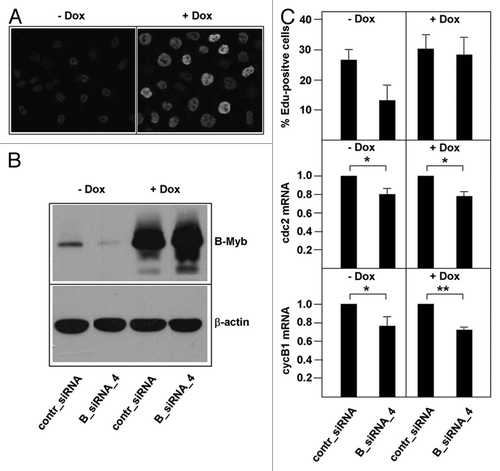
Figure 5. Interaction of B-Myb and Pdip1. (A and B). QT6 cells were transfected with expression vectors for B-Myb, c-Myb and Myc-Pdip1, as indicated at the bottom. After 16 h, the cells were radiolabeled with 35S-methionine and cell extracts were subsequently immunoprecipitated with antibodies against B-Myb, c-Myb or the Myc-tag. Immunoprecipitates were then analyzed by SDS-PAGE and autoradiography. Protein bands corresponding to B-Myb, c-Myb and Pdip1 are marked. (C). Co-immunoprecipitation of endogenous B-Myb and Pdip1. Cell extract from HEK293 cells was precipitated with rabbit antiserum against Pdip1 or an unrelated protein (control IgG). The immunoprecipitates and an aliquot (5%) of the total-cell extract (TCE) were analyzed by western blotting with antibodies against B-Myb. The asterisk marks a non-specific protein band. The bottom part of (C) shows a sample of the total-cell extract stained with the Pdip1-specific antiserum.
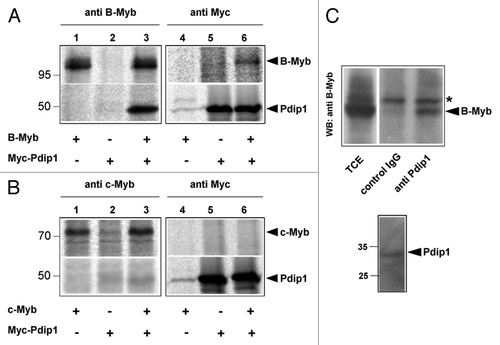
Figure 6. Mapping the binding region for B-Myb within Pdip1. Top: Schematic illustration of Pdip1 deletion constructs. The BTB-domain is highlighted. Bottom: QT6 cells were transfected with expression vectors for B-Myb and the indicated Myc-tagged Pdip1 constructs. Cells were then analyzed by immunoprecipitation as in . Protein bands corresponding to B-Myb and Pdip1 constructs are marked.
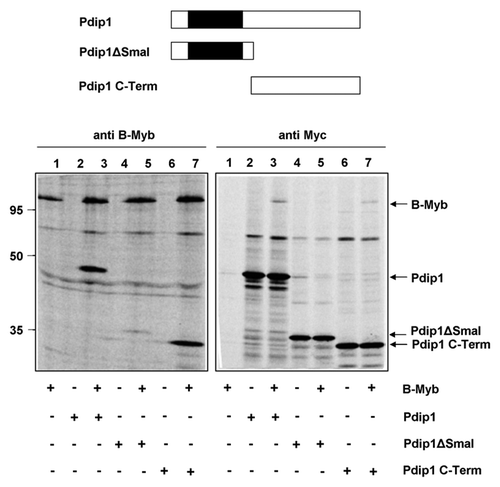
Figure 7. Mapping the binding region for Pdip1 within B-Myb. (A). Schematic illustration of B-Myb constructs. R1, R2, R3 and TAD refer to the repeats of the Myb DNA-binding domain and the transactivation domain, respectively. Small numbers refer to amino acids. c-Myb sequences are represented by black bars. (B and C). QT6 cells were transfected with expression vectors for Myc-Pdip1 and the indicated B-Myb constructs. Cells were radiolabeled with 35S-methionine and analyzed by immunoprecipitation with antibodies against B-Myb (top panels) or the Myc-tag (bottom panels), as described for . In lanes 6 to 8 of (C), antiserum against c-Myb instead of B-Myb was used. Protein bands corresponding to Pdip1 and the B-Myb constructs are marked. Pdip1 co-precipitated via B-Myb is marked with an asterisk.
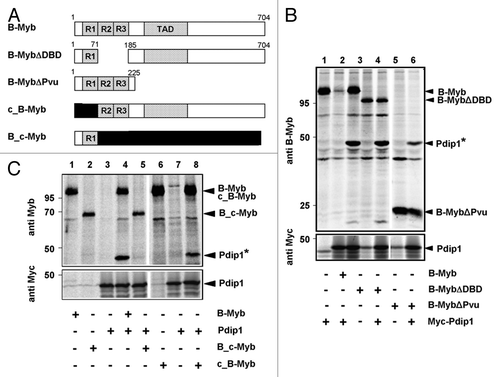
Figure 8. Pdip1 binds to sequences overlapping with the B-Myb transactivation domain. (A). Schematic illustration of GST- and YFP-B-Myb constructs. (B). GST-pull down experiments were performed with the indicated GST and GST-B-Myb fusion proteins and lysates of QT6 cells expressing Myc-Pdip1. Bound proteins (lanes 2 to 4) and an aliquot (10%) of the input sample (lane 1) were analyzed by western blotting using a Myc-specific antibody. An anti-GST western blot of the bacterial proteins used in the pull-down experiment is shown in the lower panel and demonstrates comparable protein amounts. (C). QT6 cells were transfected with the indicated combinations of plasmids encoding Myc-Pdip1, YFP-B-Myb, YFP-B-Myb D3 (345–440) and YFP. Cells were lysed 24 h post-transfection and the soluble fraction was incubated with GFP-trap beads, resulting in the binding of YFP-fusion proteins and their binding partners to the beads. Bound proteins and aliquots (6%) of the input extracts were analyzed by western blotting with antibodies against Myc and GFP.
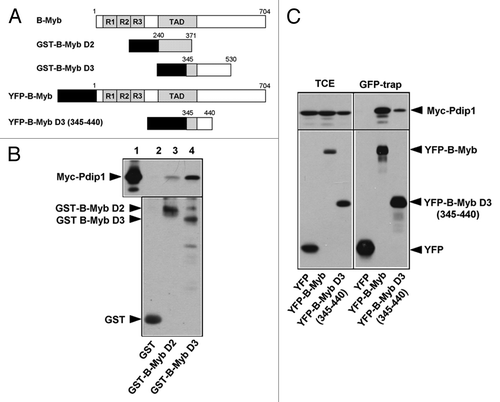
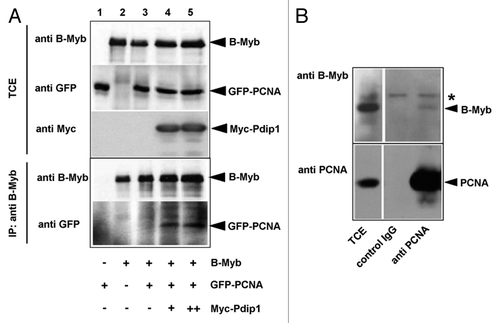
Figure 9. Co-immunoprecipitation of B-Myb and PCNA. (A). QT6 cells were transfected with expression vectors for B-Myb, GFP-PCNA and Myc-Pdip1, as indicated at the bottom. Cell extracts prepared 16 h after transfection were then immunoprecipitated with antibodies against B-Myb. The immunoprecipitates and aliquots (5%) of the total-cell extract (TCE) were analyzed by western blotting with antibodies against B-Myb (top panels) or GFP (bottom panels). Protein bands corresponding to B-Myb and GFP-PCNA are marked. (B). Co-immunoprecipitation of endogenous B-Myb and PCNA. Cell extracts from untransfected Hek293 cells were precipitated with antibodies against PCNA or with control IgG, as indicated. The immunoprecipitates and aliquots (2.5%) of the total-cell extracts (TCE) were analyzed by western blotting with antibodies against B-Myb. The asterisk marks a non-specific protein band.