Figures & data
Figure 1. siRNA-mediated downregulation of αSNAP alters the autophagic flux. (A) SK-CO15 cells were transfected with either control or two different αSNAP-specific siRNA duplexes (D1 and D2). Expression of αSNAP and autophagic markers, LC3, NBR1 and p62 in total-cell lysates was determined by immunoblotting. (B) siRNA depletion of αSNAP was performed in either control SK-CO15 cells (SK-neo) or cells with stable expression of siRNA-resistant bovine αSNAP (SK-αSNAP). Expression of αSNAP and autophagic markers in cell lysates was determined by immunoblotting 48 h post-transfection. (C and D) HeLa-GFP-LC3 cells were transfected with either control or αSNAP-specific siRNAs and formation of autophagosomes was visualized by confocal microscopy analysis of GFP fluorescence in fixed cells 72 h post-transfection. Data in this and other figures are presented as mean ± SEM of three independent experiments. *p < 0.001 compared with control siRNA-transfected cells. Scale bar, 20 µm.
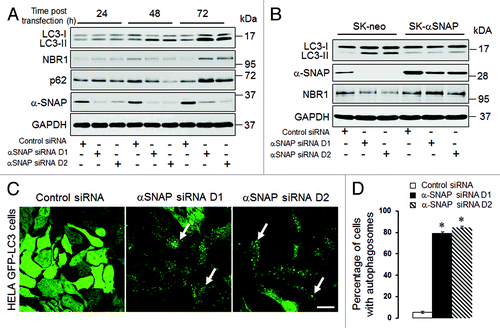
Figure 2. Lysosomal inhibitors exaggerate LC3 conjugation and accumulation of autophagosomes in αSNAP-depleted epithelial cells. SK-CO15 (A and B) or HeLa-GFP-LC3 (C) cells transfected with either control or αSNAP duplex 1 siRNAs were treated for 4 h with either vehicle or lysosomal inhibitors bafilomycin A (0.2 µM) or chloroquine (100 µM). Expression of LC3-II was determined in SK-CO15 cells by immunoblotting, whereas formation of autophagosomes was examined in HeLa-GFP-LC3 cells by fluorescence microscopy. **p < 0.05 compared with the vehicle-treated αSNAP-depleted cells. Scale bar, 20 µm.
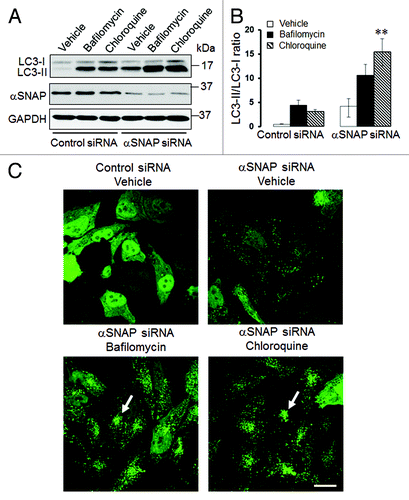
Figure 3. Loss of αSNAP decreases expression of mTOR and its downstream effector, 4E-BP1. SK-CO15 cells were transfected with either control or αSNAP-specific siRNAs. Expression and phosphorylation of mTOR and its downstream effectors p70 S6 kinase and 4E-BP1 in total-cell lysates was determined at different times post-transfection.
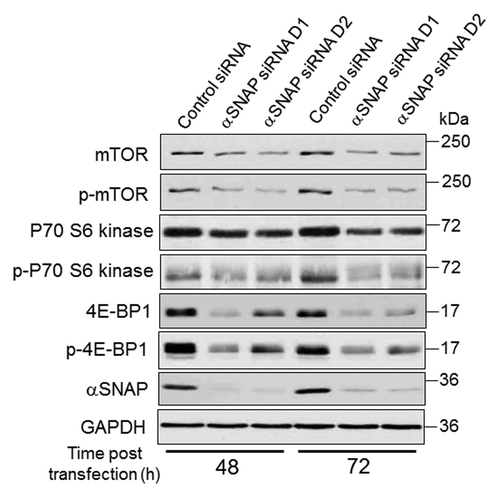
Figure 4. Atg5 and Atg7 play roles in induction of autophagy caused by downregulation of αSNAP. (A and B) SK-CO15 cells were subjected to sequential transfections with one of the following siRNA pairs: control-control, control-Atg5, control-Atg7, control-αSNAP, Atg5-αSNAP or Atg7-αSNAP. Expression of LC3, αSNAP, Atg5 and Atg7 was determined by immunoblotting 48 h after the second transfection. #p < 0.01 compared with control-αSNAP siRNA-transfected cells. (C and D) HeLa-GFP-LC3 cells were sequentially transfected with control-control, control-Atg5, control-αSNAP and Atg5-αSNAP siRNA combinations and formation of autophagosomes was analyzed by fluorescence spectroscopy at 72 h after the second transfection. *p < 0.001 compared with control-αSNAP siRNA-transfected cells. Scale bar, 20 µm.
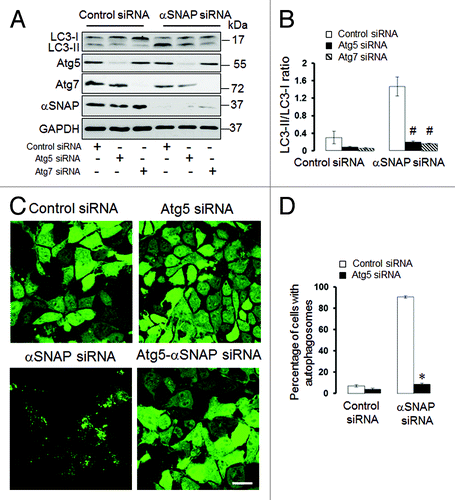
Figure 5. Loss of αSNAP triggers fragmentation of the Golgi in parallel to autophagy induction. Control and αSNAP-depleted HeLa-GFP-LC3 cells were immunofluorescence labeled for Golgi markers Giantin and TGN46 (red) at 72 h post-transfection. Control cells are characterized by the compact perinuclear Golgi complex, whereas αSNAP depletion results in a dramatic fragmentation of the Golgi (arrows) and appearance of Golgi markers in LC3-positive autophagosomes (inserts). Scale bar, 10 µm.
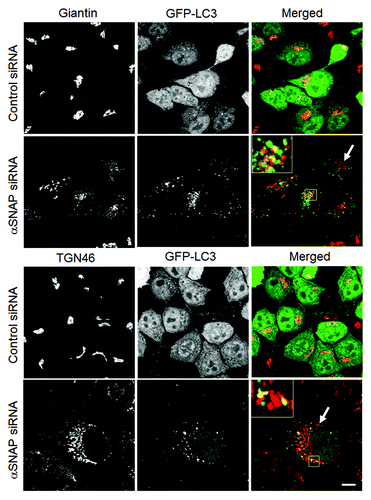
Figure 6. Pharmacological disruption of the Golgi stimulates autophagy. SK-CO15 (A and B) and HeLa-GFP-LC3 (C and D) cells were treated for 24 h with either vehicle, Brefeldin A (2 µM) or Golgicide A (50 µM), and expression of LC3-II and accumulation of autophagosomes were determined by immunoblotting and fluorescence microscopy, respectively. *p < 0.001 compared with the vehicle-treated group (n = 3). Scale bar, 20 µm.
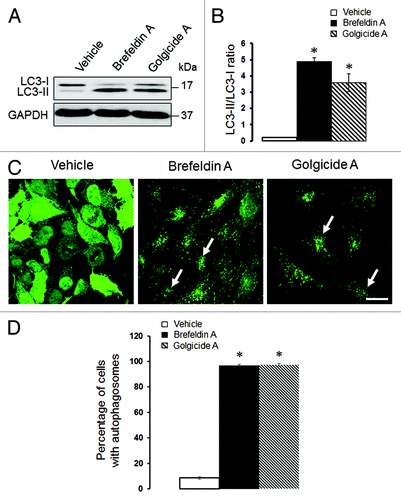
Figure 7. Loss of αSNAP decreases expression of Brefeldin-sensitive guanine nucleotide exchange factors. (A) Expression of Brefeldin-sensitive exchange factors GBF1, BIG1 and BIG2 was examined in control and αSNAP-depleted SK-CO15 cells 48 h after siRNA transfection. (B) Effect of αSNAP knockdown on localization of GBF1 (red) in SK-CO15 cells was analyzed by immunofluorescence labeling and confocal microscopy. Scale bar, 20 µm.
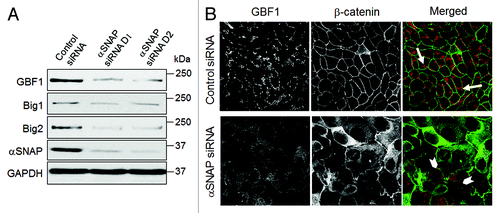
Figure 8. Depletion of GBF1, but not other Brefeldin-sensitive exchange factors, accelerates epithelial cell autophagy. (A and B) SK-CO15 cells were transfected with either control GBF1 or a combination of BIG1 and BIG2 siRNAs, and expression of targeted proteins and LC3 was examined at two different times post-transfection. (C and D) HeLa-GFP-LC3 cells were transfected with either control GBF1 or BIG1, plus BIG2 siRNAs, and formation of autophagosomes was analyzed by fluorescence microscopy. *p < 0.001 compared with control siRNA-transfected cells. Scale bar, 20 µm.
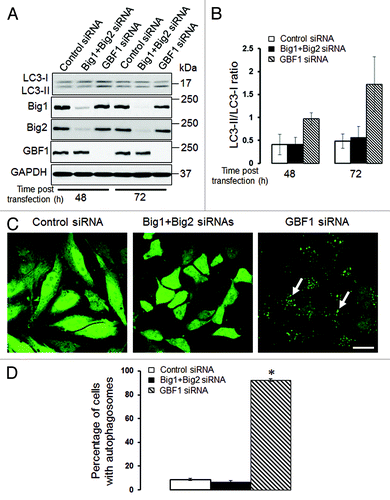
Figure 9. Bif-1 is involved in the enhanced autophagy caused by downregulation of αSNAP. SK-CO15 (A and B) and HeLa-GFP-LC3 cells were subjected to sequential transfections with one of the following siRNA pairs: control-control, control-Bif-1, control-αSNAP and Bif-1-αSNAP. Levels of LC3 and Bif-1, as well as accumulation of autophagosomes, were determined by immunoblotting and fluorescence microscopy, respectively. #p < 0.01 compared with the control-αSNAP siRNA-transfected group. Scale bar, 20 µm.
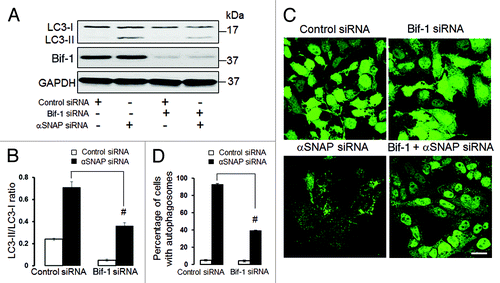
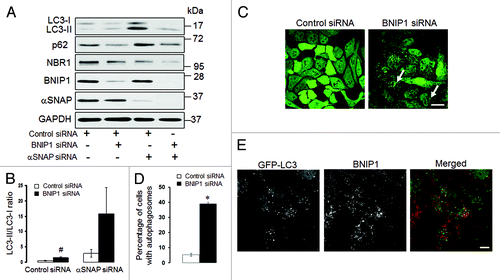
Figure 10. Enhanced autophagy in αSNAP-depleted epithelial cells does not depend on BNIP1. (A and B) SK-CO15 cells were subjected to sequential transfections with one of the following siRNA pairs: control-control, control-BNIP1, control-αSNAP and BNIP1-αSNAP. Expression of targeted proteins and autophagic markers was determined by immunoblotting at 48 h after the second transfection. #p < 0.01 compared with control siRNA-transfected cells. (C and D) HeLa-GFP-LC3 cells were transfected with either control or BNIP1-specific siRNAs and accumulation of autophagosomes was monitored by fluorescence spectroscopy. Scale bar, 20 µm; *p < 0.001 compared with control siRNA-transfected cells. (E) HeLa-GFP-LC3 cells subjected to αSNAP depletion were immunolabeled for BNIP1 (red) at 72 h post-transfection. Scale bar, 10 µm.