Figures & data
Figure 1. Expression of FoxO1 and Aurora A kinase is inversely related. (A) Western blot analysis of total FoxO1, phospho-FoxO1 (S256), p21 and Aurora A expression at different cell cycle stages in HepG2 cells. Equal loading of protein was confirmed by GAPDH. (B) Quantitative real-time PCR of FoxO1 and Aurora A expression at different cell cycle stages of HepG2 cells. RNA levels were normalized to GAPDH. (C) Cell cycle phases of HepG2 cells were confirmed by FACA analysis. (D) Knockdown of Aurora A was confirmed by quantitative real-time RT-PCR. The graph represents means ± SD for the change in the mRNA level relative to GAPDH from three independent experiments. (E) Expression of FoxO1 and FoxO3a in Aurora A-knockdown cells was measured by quantitative real-time RT-PCR. GAPDH expression was examined as an internal control. Data represent the means ± SD of three independent experiments.
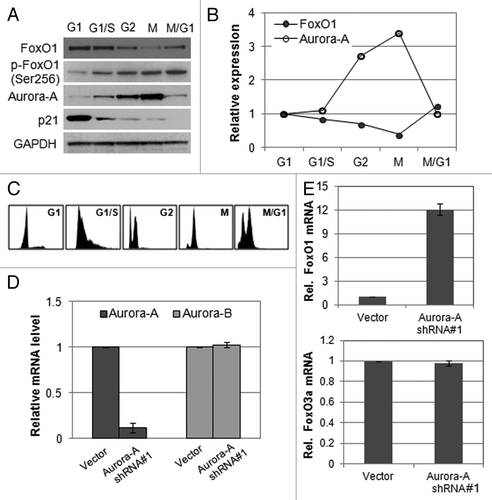
Figure 2. Depletion of Aurora A induces upregulation of FoxO1 expression in HepG2 and SK-HEP-1 cells, and RNAi-resistant Aurora A downregulates FoxO1 expression. (A) Western blot analysis of total FoxO1, phospho-FoxO1 (S256), p21 and Aurora A expression in vector or Aurora A shRNA-expressing HepG2 and SK-HEP-1 cells. (B) Western blot analysis of total FoxO1, phospho-FoxO1 (S256), p21 and Aurora A expression in vector or Aurora A shRNA-expressing Huh-7 and Hep3B cells. Two different Aurora A shRNA were used. Equal loading of protein was confirmed by GAPDH. (C) Nucleotides substituted (base changes are underlined) in the vector expressing the RNAi-resistant Aurora A. (D) HepG2 cells were transduced with pLKO.1 or Aurora A shRNA lentiviral particles. At 48 h after the transduction, cells were transfected with RNAi-resistant Aurora A containing or empty vector. Protein lysates were extracted 48 h later and followed by western blot analysis with the indicated antibodies. (E) Cell proliferation in control and Aurora A-knockdown cells that expressed either empty vector or RNAi resistant Aurora A were measured on the indicated days. Data represent the means ± SD of three independent experiments.
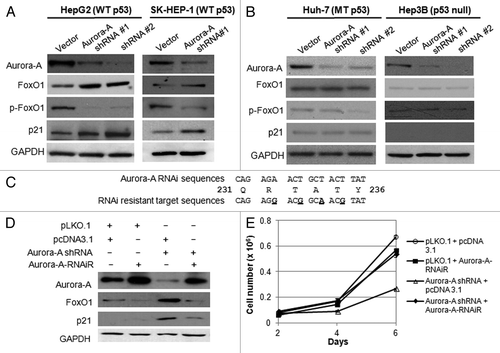
Figure 3. Silencing of Aurora A induces G2/M arrest. (A) The growth of vector and Aurora A shRNA HepG2 cells were measured on the indicated days. Data shown are the mean ± SD of three independent experiments. (B) Cell cycle profile of synchronized vector control and Aurora A shRNA HepG1, Huh-7 and Hep3B cells was analyzed by flow cytometry analysis. Percent of G2/M cells is shown. (C) The level of FoxO1, cyclin B1 and Aurora A during M phase was measured by western blot analysis. Nocodazole (100 ng/ml) arrested control and Aurora A shRNA HepG2 and Hep3B cells were released from nocodazole block, and protein lysates were prepared at the indicated time thereafter. Equal loading of protein was confirmed by GAPDH.
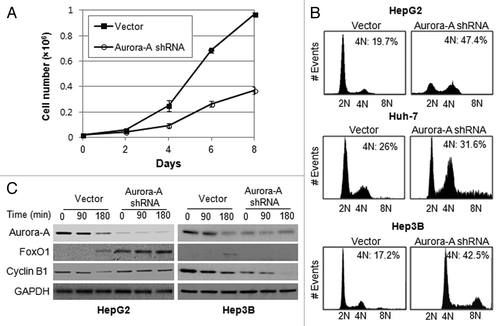
Figure 4. Depletion of FoxO1 in Aurora A-knockdown cells lead to exit G2/M arrest and induce apoptosis. (A) FoxO1-specific siRNA was transfected into vector control and Aurora A shRNA HepG2 cells, and knockdown of FoxO1 was confirmed by western blot analysis. (B) Cell cycle profiles in scrambled or FoxO1-specific siRNA-transfected control and Aurora A-knockdown HepG2 cells were monitored at 72 h after transfection by flow cytometry analysis. Representative profiles and percent of subG1, G0/G1, S, G2/M cells from one of two independent experiments are shown. (C) The percentages of TUNEL-positive nuclei in scrambled or FoxO1-specific siRNA transfected control and Aurora A-knockdown cells 96 h after transfection. Data represent the means ± SD of two independent experiments.
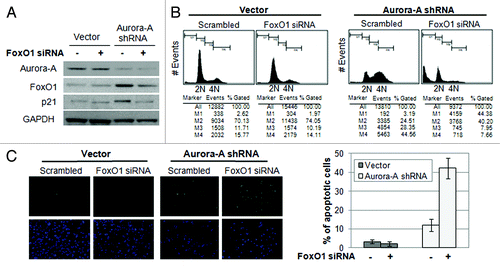
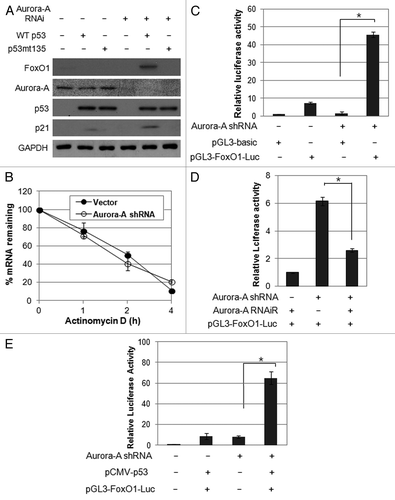
Figure 5. Aurora A regulates FoxO1 expression at the transcriptional level in a p53-dependent manner. (A) Hep3B cells were co-transfected with a combination of indicated plasmids, incubated for 48 h, and western blot analysis was performed. (B) FoxO1 mRNA half-life was determined by actinomycin D treatment. Vector control and Aurora A shRNA cells were treated with actinomycin D (10 μg/ml) for the indicated time points. FoxO1 mRNA levels were measured by real-time qRT-PCR. (C) For luciferase reporter assay, vector control and Aurora A-knockdown cells were transfected with luciferase reporter plasmid containing FoxO1 promoter. pGL3 vector was used as a negative control. Data shown are mean ± SD of three independent experiments. *p < 0.01. (D) HepG2 cells were transfected with a combination of the Aurora A shRNA and RNAi-resistant Aurora A variant in the presence of luciferase reporter plasmid containing FoxO1 promoter. pGL3 vector was used as a negative control. Data shown are mean ± SD of three independent experiments. *p < 0.05. (E) Hep3B cells were co-transfected with a combination of the indicated plasmids in the presence of luciferase reporter plasmid containing FoxO1 promoter. pGL3 vector was used as a negative control. Data shown are mean ± SD of three independent experiments. *p < 0.05.