Figures & data
Figure 1. The SCF complex is required for Ku80 ubiquitylation and degradation in response to DSBs. (A) 35S-labeled Ku80 was added to extract that was either immunodepleted of Cul1, mock-depleted, or mock-depleted and contained GST-tagged dominant-negative Cul1. At one hour intervals, protein was precipitated using trichloro-acetic acid and quantified using a scintillation counter. Average and standard deviation of three independent experiments at each time point are shown. (B) Extracts containing either GST or GST-DN-Cul1 were incubated with SB-DNA beads and inputs and co-purifying proteins were probed with antibodies against Ku80, Skp1, and H3. Beads from a duplicate experiment were probed with antibodies against Cul1, Skp1, and H3 (a loading control). (C) 35S-labeled Ku80 and GFP-SBP, as a loading control, were added to egg extract. After a 45 min pre-incubation, either GST or GST-DN-Cul1 was added. The extract was then incubated with SB-DNA beads. After 30 min, beads were removed from the extract and washed once with and added to extract containing GST or GST-DN-Cul1 but lacking labeled Ku80. Beads were incubated in this extract for an additional 0 (t = 30), 30 (t = 60), or 60 (t = 90) minutes. Bead fractions were visualized by phosphorimager. (D) Ku80 remaining on beads was quantified using a phosphorimager after normalizing to GFP-SBP in three independent experiments. Error bars indicate one standard deviation.
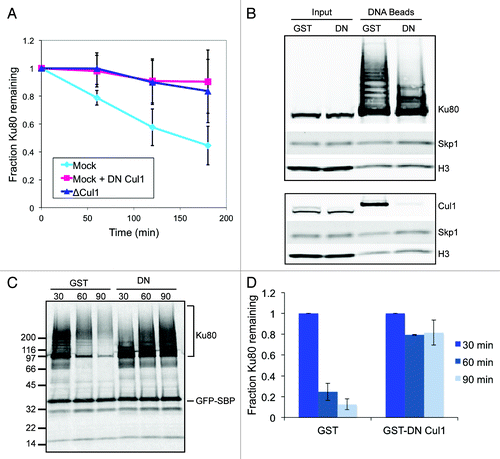
Figure 4. Fbxl12 is required for SCF interaction with DSBs and Ku80 ubiquitylation. Fbxl12 was immunodepleted from egg extracts, which were then incubated with SB-DNA beads. Interacting proteins were analyzed by immunoblot. The arrow points to a Nedd8-conjugated Cul1, which can be detected by anti-ubiquitin antibody. A likely cross-reacting band, which is not depleted by anti-Fbxl12 antibodies, is indicated with an asterisk.
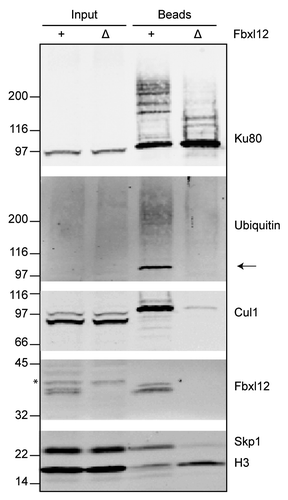
Table 1. Xenopus F box proteins were screened for Ku sensitivity
Figure 2. The F-box protein Fbxl12 binds to DSBs in a Ku-sensitive manner. (A) 35S-labeled Xenopus tropicalis Fbxl12 and histone H3 were added to egg extract that was either mock- or Ku-depleted. Streptavidin-coated (SA), SB-DNA, or DB-DNA beads were added to the mock-depleted extract and SB-DNA beads were added to the Ku-depleted extract. Bead fractions were visualized by phosphorimager. (B) Protein alignment of Fbxl12 amino acid sequence from Xenopus laevis, Xenopus tropicalis, Homo sapiens and Mus musculus. Indicated are the F box (blue box), conserved F box residues that were mutated (see , red boxes), and the start of the ∆N-Fbxl12 truncation mutant. (C) Egg extracts were either mock- or Ku-depleted, and 1 µM recombinant MBP-Fbxl12 (MF) was then added. These extracts were then incubated with SB-DNA beads, and co-purifying proteins were probed with antibodies against MBP, Ku70, Skp1 or H3. (D) Mock- or Ku-depleted extract was incubated with SB-DNA beads, and bound proteins were probed using antibodies against Xenopus laevis FBXL12. A likely cross-reacting band is indicated with an asterisk.
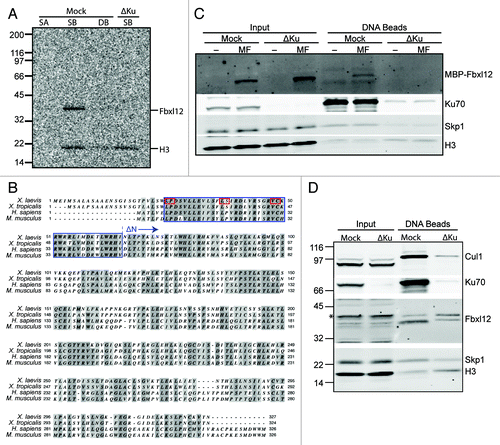
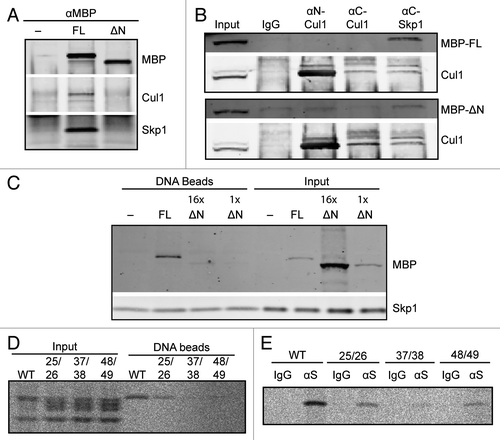
Figure 3. The FBXL12 F-box is required for interaction with SCF components and with DSBs. (A) Extracts containing buffer, MBP-Fbxl12 (FL), or MBP-∆N-Fbxl12 (∆N) were immunopurified using an anti-MBP antibody. Copurified proteins were probed by immunoblot. (B) MBP-Fbxl12 (MBP-FL) or MBP-∆N-Fbxl12 (MBP-∆N) were added to extract, which was then immunpurified using antibodies against N-terminal Cul1, C-terminal Cul1, or C-terminal Skp1. Co-purified proteins were probed using antibodies against MBP and Cul1. (C) MBP-Fbxl12 (FL) was added to egg extract for a final concentration of 1 µM, and MBP-∆N-Fbxl12 (∆N) for a final concentration of 1 ∆M (1x) or 16 ∆M (16x). Extracts were then incubated with SB-DNA beads, and bound proteins were analyzed by immunoblot. (D–E) 35S-labeled Fbxl12 containing the mutations L25A, P26A (25/26), L37A, S38A (37/28), or V48A, C49A (48/49) or wild type Fbxl12 (WT) was added to egg extract and allowed to bind SB-DNA beads (D) or either IgG or anti-Skp1 antibodies (∆S) (E). Bead fractions were analyzed by phosphorimager.