Figures & data
Figure 1. Mph1 is required for the proper segregation of recombined homologous chromosomes during meiosis I. (A) The meiotic segregation of chromosome II was scored in a wild-type h90 cen2-GFP strain (JG12618) and an h90 cen2-GFP strain carrying the knockout allele of mph1 (mph1Δ) (JG15607). Cells were stained with Hoechst and examined under the fluorescence microscope. Chromosome segregation was scored in at least 100 asci. (B) The strains described in (A) were fixed and immunostained for tubulin and GFP. DNA was visualized by Hoechst staining. A total of 100 anaphase I cells were examined under a fluorescence microscope, and the segregation of chromosome II, marked by cen2-GFP, was scored.
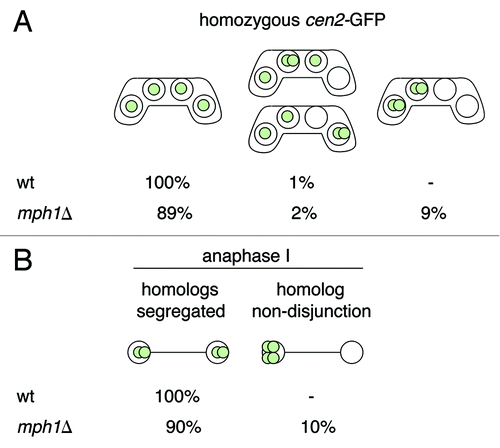
Figure 2. Segregation of sister centromeres in spo4Δ and spo6Δ meiotic cells. (A) The wild-type strain h− lys1-GFP (wt) (JG11338) and h− lys1-GFP strains carrying knockout alleles of spo4 (spo4Δ) (JG14885) or spo6 (spo6Δ) (JG14888) were crossed to h+ strains of the same genotype but lacking lys1-GFP (JG11339, JG14872 and JG14879, respectively). Similarly, strains carrying a knockout allele of spo4 transformed with a plasmid carrying either a wild-type allele of spo4 (spo4Δ spo4+) (JG14911) or a “kinase-dead” allele of spo4 (spo4Δ spo4K95A) (JG14913) were crossed to h+ strains of the same genotype but lacking lys1-GFP (JG14903 and JG14907, respectively). Cells were sporulated and stained with Hoechst. Segregation of chromosome I was scored in at least 100 asci. (B) The strains described in (A) were fixed and stained with antibodies against tubulin and GFP. DNA was visualized by Hoechst staining. Cells were examined under a fluorescence microscope and segregation of chromosome I, marked by lys1-GFP, was scored in 100 anaphase I or anaphase II cells.
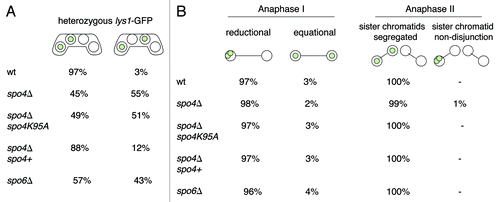
Figure 3. The anaphase II spindles are abnormally expanded in spo4Δ and spo6Δ mutant cells. A wild-type h90 strain and h90 strains carrying either a knockout allele of spo4 (spo4Δ) (JG14875), or a knockout allele of spo6 (spo6Δ) (JG14882) or a knockout allele of spo4 transformed with a plasmid carrying either a wild-type allele of spo4 (spo4Δ spo4+) (JG14906) or a “kinase-dead” allele of spo4 (spo4Δ spo4K95A) (JG14910) were sporulated, fixed and stained with antibodies against tubulin and GFP. DNA was visualized by Hoechst staining. The length of meiosis II spindles was determined in 100 zygotes.
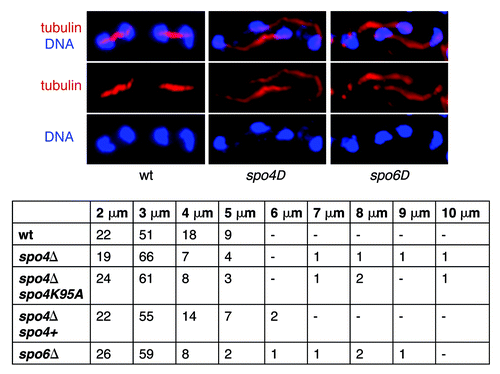
Figure 4. Live-cell analysis of anaphase II spindles in spo4∆ cells. The wild-type strain h− mCherry-atb2 hta2-TagBFP (wt) (JG16499) (A) or the h− mCherry-atb2 hta2-BFP strain carrying a knockout allele of spo4 (spo4Δ) (JG16662) (B) was crossed to h+ strains of the same genotype (JG16486 and JG16663, respectively). Cells were sporulated and spindle elongation during meiosis II was analyzed by live cell imaging. The numbers indicate time in minutes. Anaphase II spindles were analyzed in four spo4Δ and eight wild-type cells.
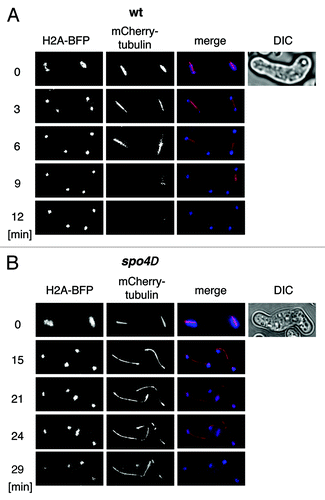
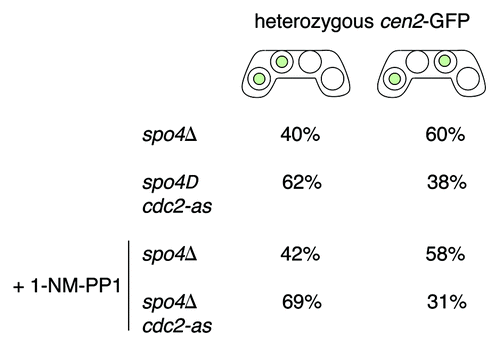
Figure 5. Inactivation of Cdc2-as partially suppresses the mutant phenotype of spo4Δ cells. The h− cen2-GFP strain carrying either a knockout allele of spo4 (spo4Δ) (JG14873) or a knockout allele of spo4 and an analog-sensitive allele of cdc2 (spo4Δ cdc2-as) (JG16848) were crossed to h+ strains of the same genotype but lacking cen2-GFP (JG14872 and JG16858, respectively) and plated on PMG-N plates. After 24–43 h of incubation at 25°C cells were washed with water and incubated in a liquid PMG-N medium with or without inhibitor (5µM 1-NM-PP1) at 25°C for 4–7 h. Cells were stained with Hoechst to visualize DNA and segregation of cen2-GFP was scored in at least 50 asci.