Figures & data
Figure 1. Targeted inactivation of Ctnnbl1 results in mid-term embryonic lethality. (A) Targeting the mouse Ctnnbl1 locus. The top line depicts the mouse Ctnnbl1 locus (three exons: E1, E2 and E3 are depicted) aligned with the targeting construct which contains a promoterless β-galactosidase/pgk-neo cassette flanked by flippase recognition sites (oval shapes) integrated into intron 2 with LoxP sites (open triangles) flanking E3. The left and right homology arms are 4.6 and 4.8 kb, respectively. Restriction endonuclease sites Ase I (A) and Sap I (S) are indicated as are the locations of the left- and right-hand probes (LHP and RHP) used for the Southern blot analysis. (B) Southern blot of tail DNA from mice carrying the targeted β-gal/neo cassette insertion into Ctnnbl1 E3 on one allele as well as controls hybridized with probes LHP and RHP. (C) Genotypes of weaned animals born to Ctnnnbl1+/− intercrosses. A similar analysis is shown for the progeny born to intercrosses of AX0016 mice that carry a gene trap insertion into the first Ctnnbl1 intron. (D) Genotypes of embryos obtained at day 8.5 or 11.5 of gestation from Ctnnnbl1+/− intercrosses. (E) Broad expression of the Ctnnbl1 locus as revealed by staining of a day 14.5 Ctnnnbl1+/− embryo for β-galactosidase activity.
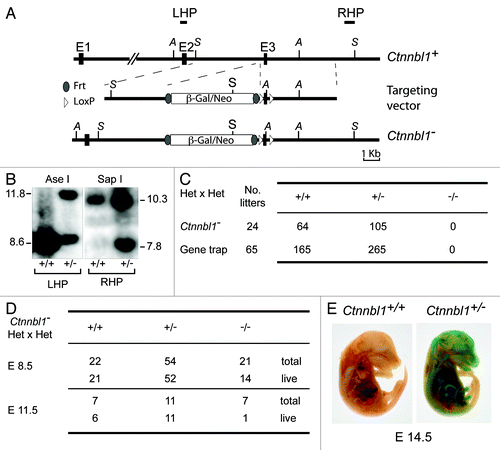
Figure 2. Lineage-specific ablation of Ctnnbl1 has little effect on B cell development. (A) B cell-specific inactivation of Ctnnbl1. The top two lines depict the wild type locus and the derivative containing a targeted β-gal/neo insertion into intron 2 as in . In alignment with these are depicted the loci obtained following (i) Cre-mediated excision of E3 (Ctnnnbl1-ΔE3; which will be inactive); (ii) following Flippase-mediated excision of the β-gal/neo cassette (Ctnnnbl1flx; which will be functional but a ‘floxable’ substrate for Cre-mediated inactivation) and (iii) both Flippase – and cre-mediated excision (yielding Ctnnnbl1ΔE3; which will be inactive). (B) Southern blot analysis of DNA extracted from sorted splenic CD43- cells (which comprise > 95% resting B cells) as well as CD43+ cells (which comprise T cells, granulocytes/macrophages and activated B cells) from mice of the indicated Ctnnbl1 genotypes that also express Cre recombinase under control of the B-cell-specific mb1 promoter. Ase I-digested DNA was hybridized with a Ctnnbl1 E2 probe. The origins of the various hybridizing bands are indicated with the results revealing highly (> 95%) efficient deletion of Ctnnbl1 E3 in the resting B cells of mb1-Cre Ctnnbl1-/flx mice. Some deletion of Ctnnbl1 E3 is also observed within the CD43+ fraction, which probably reflects activated B cells within this population. (C) Loss of CTNNBL1 expression in the sorted splenic B cells of mb1-Cre Ctnnbl1-/flx mice as judged by (i) RT-PCR analysis of RNA using primers specific for Ctnnbl1 E3 relating to HPRT (as a control) and (ii) western blot analysis of protein using α-tubulin as a control. (D) Comparison of splenic B and T cell populations in mb1-Cre Ctnnbl1-/flx mice (squares) as compared with mb1-Cre Ctnnbl1+/flx controls (circles). T cells were identified as CD3+ and subdivided into CD4+ and CD8+ single-positive subpopulations. B cells were identified as B2220+ or CD19+ with B220+ CD19+ cells divided into B1 (CD23- CD21-); follicular (FO: CD23high CD21+) or marginal zone (MZ: CD23low CD21+) subpopulations. (E) Comparison of pre- and pro- B cell populations in the bone marrow of mb1-Cre Ctnnbl1-/flx mice (squares) as compared with mb1-Cre Ctnnbl1+/flx controls (circles). Pre-B cells were defined as CD43- B220+ IgM- whereas pro-B cells are as CD43- B220+ IgM+.
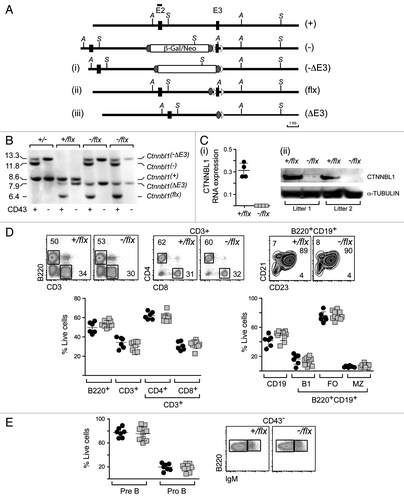
Figure 3. CTNNBL1-deficient B cells give diminished immunoglobulin class-switch recombination. (A) Flow cytometric analysis of switching to IgG1 in spleen cells from mb1-Cre Ctnnbl1-/flx and control mice that had been cultured for 3 d in the presence of LPS+IL4. (i) Dot plots depict results from two pairs of littermates in independent experiments. (ii) Bar graphs on the right present the means ± sd from analysis of multiple mice as indicated. The values within each experiment are normalized to the average value of the control samples. (B) Abundance of AID in the day 3 LPS+IL4-activated B cells obtained from two littermate pairs of mb1-Cre Ctnnbl1+/flx and mb1-Cre Ctnnbl1-/flx mice analyzed by western blot using α-tubulin as a control. (C) Analysis of switching to IgG1 as a function of B cell clonal expansion in CFSE-labeled splenic cells from mb1-Cre Ctnnbl1-/flx and control mice that had been cultured for 3 d in the presence of LPS+IL4. (i) The CFSE-staining profiles for two pairs of litter-matched mice with the number of cell divisions undergone (inferred from dilution of the CFSE label) indicated. (ii) The percentage of cells that have switched to IgG1 as a function of number of cell divisions undergone in six independent experiments.
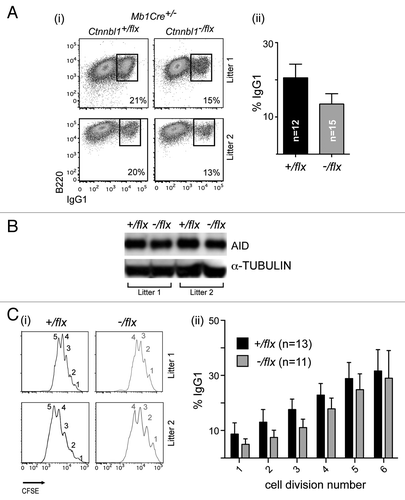
Figure 4. CTNNBL1-deficient B cells are slow to enter their first S-phase on LPS activation. (A) Clonal expansion of splenic B cells from three littermate pairs of mb1-Cre Ctnnbl1-/flx and control mice as monitored by CFSE dilution. (B) Cell cycle analysis of splenic B cells from multiple littermate pairs of mb1-Cre Ctnnbl1-/flx and control mice as analyzed at 24 and 48 h of incubation with LPS/IL4. (i) The propidium iodide staining profiles of the cells from two littermate pairs are shown on the left with (ii) the results from multiple animals summarized on the right. The proportion of cells per stage of the cell cycle is normalized to the average number of cells in the controls within each independent experiment (bars indicate mean and sd). (C) DNA synthesis in splenic B cells from littermate pairs of mb1-Cre Ctnnbl1-/flx and control mice cultured for the indicated lengths of time with bromodeoxyuridine (BrdU) and LPS. (i) Cells were analyzed for BrdU content by staining with anti-BrdU antibody and total DNA content by propidium iodide staining. The individual staining profiles are indicated (percent of cells in G0/G1 and S phase in black, with the ratio of cells in G1 vs. S that have entered a second cell division in gray) with (ii) a separate graph showing the percentage of cells that have incorporated BrdU at the various time points in four pairs of littermates.
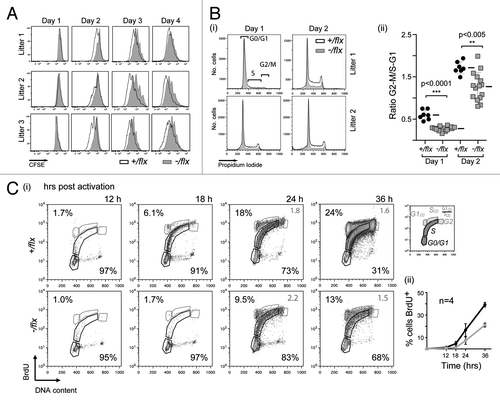
Figure 5. CTNNBL1 deficiency delays cell enlargement and S-phase entry but not the upregulation of early activation markers. (A) Comparison of blasting of splenic B cells from littermate pairs of mb1-Cre Ctnnbl1-/flx and control mice after 24 h of incubation with LPS as monitored by cell scatter analysis. (i) Individual contour plots of live cells from two of the littermates pairs with the gating for blasts vs. resting cells indicated. (ii) Percentage of large cells (blasts) gate in multiple B cell cultures 24 h post activation (mean and sd are indicated). (B) Induction of expression of cyclin D3, CDK6 and phosphorylated Rb after 24 h of incubation with LPS as monitored by western blot analysis. The abundance of lamin and α-tubulin served as loading controls. (C) Surface expression of CD69, CD86 and MHC class II on splenic B cells from littermate pairs of mb1-Cre Ctnnbl1-/flx and mb1-Cre Ctnnbl1+/flx control mice as analyzed after various times of incubation with LPS. (i) Histogram plots from a representative experiment and (ii) line graphs depicting the median fluorescence intensity at each time point derived from eight experiments. (Averages and sds are indicated). (D) Comparison of the blasting of splenic follicular and marginal zone B cells following LPS activation. The B cells were obtained from CD19-Cre Ctnnbl1-/flx and CD19-Cre Ctnnbl1+/flx control mice [with the CD19-Cre giving, like the mb1-Cre, efficient B cell-specific deletion of Ctnnbl1 (Fig. S1)]. (i) Histogram plots depicting the electronic volumes of sorted follicular and mantle zone B cells. The purity (assessed by flow cytometry of surface markers) of the sorted populations at the start of the cultures is indicated. (ii) The proportion of marginal zone B cells with diameter > 9 µ at different times post stimulation in multiple samples (means and sem shown).
![Figure 5. CTNNBL1 deficiency delays cell enlargement and S-phase entry but not the upregulation of early activation markers. (A) Comparison of blasting of splenic B cells from littermate pairs of mb1-Cre Ctnnbl1-/flx and control mice after 24 h of incubation with LPS as monitored by cell scatter analysis. (i) Individual contour plots of live cells from two of the littermates pairs with the gating for blasts vs. resting cells indicated. (ii) Percentage of large cells (blasts) gate in multiple B cell cultures 24 h post activation (mean and sd are indicated). (B) Induction of expression of cyclin D3, CDK6 and phosphorylated Rb after 24 h of incubation with LPS as monitored by western blot analysis. The abundance of lamin and α-tubulin served as loading controls. (C) Surface expression of CD69, CD86 and MHC class II on splenic B cells from littermate pairs of mb1-Cre Ctnnbl1-/flx and mb1-Cre Ctnnbl1+/flx control mice as analyzed after various times of incubation with LPS. (i) Histogram plots from a representative experiment and (ii) line graphs depicting the median fluorescence intensity at each time point derived from eight experiments. (Averages and sds are indicated). (D) Comparison of the blasting of splenic follicular and marginal zone B cells following LPS activation. The B cells were obtained from CD19-Cre Ctnnbl1-/flx and CD19-Cre Ctnnbl1+/flx control mice [with the CD19-Cre giving, like the mb1-Cre, efficient B cell-specific deletion of Ctnnbl1 (Fig. S1)]. (i) Histogram plots depicting the electronic volumes of sorted follicular and mantle zone B cells. The purity (assessed by flow cytometry of surface markers) of the sorted populations at the start of the cultures is indicated. (ii) The proportion of marginal zone B cells with diameter > 9 µ at different times post stimulation in multiple samples (means and sem shown).](/cms/asset/36716857-d1c9-4d08-8ff4-e7d0994e2b61/kccy_a_10923594_f0005.gif)
Figure 6. Comparison of RNA splicing in CTNNBL1-proficient and –deficient B cells. (A) Box Whisker plots depicting the distribution of splice indices defined as the ratio of the log2-transformed density of exonic vs. intronic reads per gene. The data include only expressed genes (defined as genes with exon/intron read densities ≥ 1). The boxes indicate the 25th to 75th percentile range with the line across the middle indicating the median and the lines above and below the box indicating the 1st and 99th percentiles. (B) Comparison of the splicing indices of individual expressed transcripts in (i) CTNNBL1-proficient vs. CTNNBL1-deficient B cells in both pre-activation (left) and post-activation (right) samples; (ii) Activated vs. resting B cells in both CTNNBL1-proficient (left) and CTNNBL1-deficient (right) samples. Whereas CTNNBL1 deficiency has no clear effect on individual splicing indices in either resting or activated samples (the correlation between the two genotypes in both sets is > 0.95), cell activation results in many genes giving altered splice indices (R < 0.79). Data on the total expression (as opposed to splicing index) of individual genes in the samples analyzed is provided in Table S1.
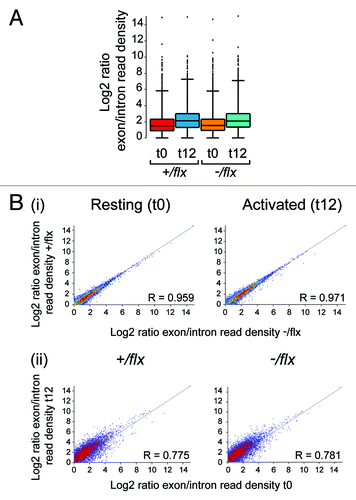
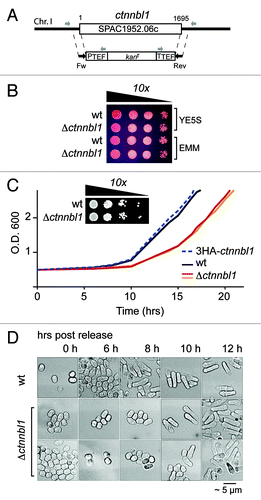
Figure 7. Loss of CTNNBL1 delays quiescence exit in S. pombe. (A) Targeting the yeast SPAC1952.06c (ctnnbl1) locus. The SPAC1952.06c gene was replaced by homologous recombination with a cassette comprising a kanamycin resistance marker under the control of translation elongation factor 1A regulatory signals as described by Bahler et al. (1998). Correct targeting was confirmed by PCR using primers whose locations are indicated. (B) Δctnnbl1 S. pombe grow similarly to controls after spotting on to YES plates. Viability of cells in log-phase growth in either yeast extract (YE) or minimal medium (EMM) from serial 10-fold dilutions spotted onto YE plates containing 5 mg/ml phloxine B. (C) Nitrogen-starved Δctnnbl1 S. pombe showed delayed initiation of growth compared with controls after transfer into rich medium. Growth curves after release from nitrogen starvation (time zero) are shown for wild type S. pombe (wt; black line), two-independent Δctnnbl1 mutants (red and orange lines) in which the ctnnbl1 locus had been replaced by a kanr cassette as well as of S. pombe carrying a control ctnnbl1 targeting in which the ctnnbl1 locus had been replaced by 3HA-tagged CTNNBL1 driven from the nmt1 promoter (blue dashed line). Similar results were obtained in four independent experiments. Growth of serial 10-fold dilutions of 3 week-starved S. pombe cultures on YE plates revealed that CTNNBL1 deficiency did not affect their viability. (D) Starved CTNNBL1-deficient S. pombe exhibit delayed exit from G0 following release from nitrogen starvation. Quiescent S. pombe, which adopt a small, round shape on nitrogen starvation, elongate prior to their first cell division on release into rich medium.Citation23 Cells were fixed in 70% ethanol at the times indicated and visualized by phase-contrast microscopy.