Figures & data
Figure 1. Karyotype of the immortal tumorigenic HA1 cell line, which originated late after transfection of human kidney cells with overexpressed telomerase. The chromosomes were prepared from metaphases and labeled by hybridization with chromosome-specific colors as described in “Materials and Methods.”
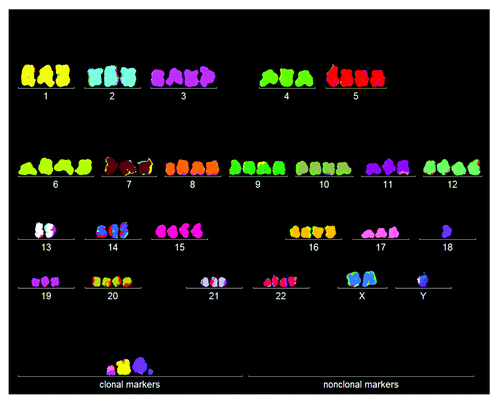
Figure 2. Three-dimensional arrays of 20 karyotypes of the immortal tumorigenic human cell lines HA1 and HA1-2 and BJ based on the cytogenetic data listed in . HA1 and HA1-2 lines originated late from human kidney cells (), and BJ from human skin cells about 60 generations after transfection of mass cultures with artificially overexpressed telomerase and two overexpressed oncogenes (text). The karyotype arrays are three-dimensional tables, which show the chromosome numbers of 20 karyotypes on the x-axis, the copy numbers of each chromosome on the y-axis and the number of karyotypes arrayed on the z-axis. By forming parallel lines, the chromosome copy numbers of 20 karyotypes indicate that the three lines are highly clonal. A minority of non-clonal copy numbers differing from the clonal values in narrow margins of ± 1, stands out above or below these lines (). This indicates that all three lines have clonal but flexible karyotypes.
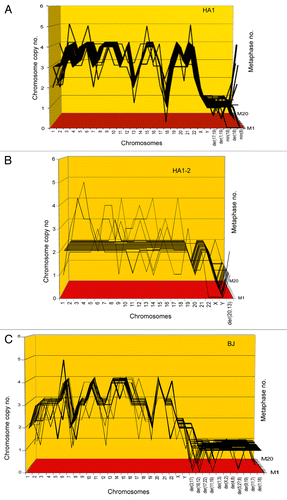
Table 1. Chromosome numbers and copy numbers of immortal, tumorigenic clones HA1, HA1–2 and BJ
Figure 3. Cellular phenotypes of human cen3tel skin cells from a centenarian female 37 (A), 100 (B), 166 (C) and 1014 (D) generations after transfection with overexpressed telomerase. The culture had a fibroblastic cellular phenotype at PD37 (A) and a less defined fibroblastic phenotype at generation PD100 (B). At generation PD166, the culture had acquired a polymorphic cellular phenotype typical of cancer cells and simultaneously became tumorigenic and immortal (C). This new polymorphic phenotype was unchanged at PD1014 (D).
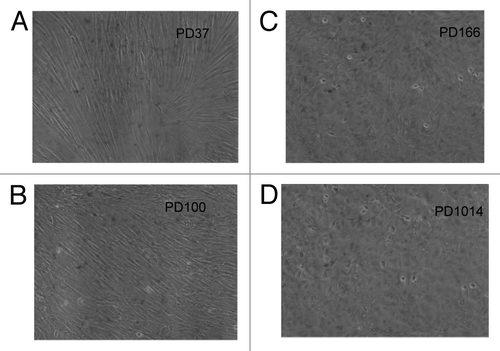
Table 2. Karyotypes of 20 cen3tel cells 37 generations after transfection with overexpressed telomerase
Table 3. Karyotypes of 20 cen3tel cells 100 generations after transfection with overexpressed telomerase
Figure 4. Aneuploidy among karyotypes of 20 human cen3tel skin cells 37 (A) and 100 (B) generations after transfection with overexpressed telomerase. The 20 karyotypes were arrayed as described for . The data show that 60% of the cen3tel cells were randomly aneuploid 37 generations after transfection (), and that 100% were randomly aneuploid 100 generations after transfection (). Moreover, the data show that the degree of aneuploidy increased sharply from generation 37 to generation 100 after transfection. The partial clonality of trisomy 7 was native of the cen3-skin biopsy studied here (see text).
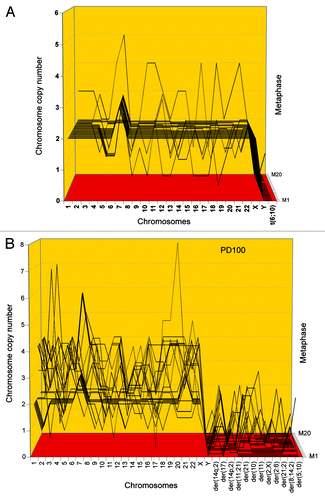
Figure 5. Karyotypic stability and drift of the tumorigenic clone cen3tel PD166 at the time of isolation (A) and after 451 (B) and 848 (C) unselected generations in vitro. As a test for immortalization, karyotype arrays prepared from the original clone cen3tel PD166 and descendents isolated 451 (PD617) and 848 (PD1014) unselected generations later were compared. These karyotype arrays show that the original cen3tel PD166 karyotype was mostly stable, but was also variable within narrow margins over 848 cell generations (). The copy numbers of 21 of 33 (64%) PD166-specific chromosomes including marker chromosomes were clone-specific during 848 generations, while 12 of 33 (36%) were generation-specific or cell-specific drifting within narrow margins of ± 1 over 848 generations ( and ). Based on the high clonal stability over 848 unselected generations, we conclude that the cen3tel PD166 clone is immortal but flexible, exceeding the operational Hayflick threshold of 50 generations by 798 clonal generations.
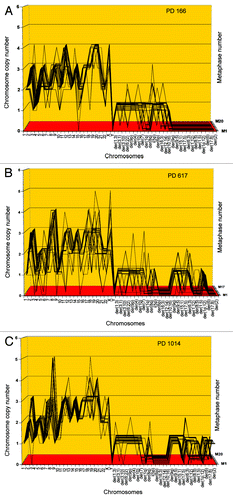
Table 4. Chromosome numbers and copy numbers of the tumorigenic clone cen3tel PD166, and at subsequent generations PD617 and PD1014
Figure 11. Karyotypic stages during carcinogenesis of a virtual organ of 106 cells: (1) The induction of random aneuploidy in an arbitrary 1% of normal cells by a carcinogen, such as overexpressed telomerase () or SV40 virus ( and ); (2) Self-catalyzed progression of aneuploidy over many cell generations up to 100% of the initiated cell population, as for example in the human skin cells 100 generations after aneuploidization by overexpressed telomerase shown in ; (3) The stochastic evolution of an autonomous cancer stem cell with a new, clonal flexible karyotype and phenotype. The new autonomous cancer cell supplants the 106 non-cancerous precursors in our Petri dish within 20 generations after its origin ( and ); (4) Self-catalyzed karyotypic variations of cancer cells generate new unselected subspecies, spontaneously () and drug-resistant or metastatic subspecies, selectively (see text).
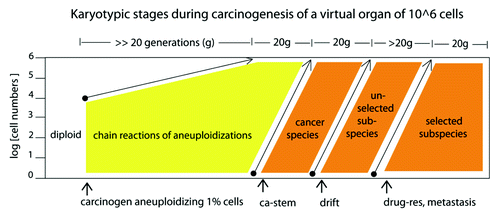
Figure 6. Two individual karyotypes of the cen3tel clone PD166 at its origin (A) and after 848 generations of unselected growth in culture at PD1014 (B). About 64% of the chromosomes of the two karyotypes share PD166-specific copy numbers or are common markers ( and ). The two karyotypes differ in generation-specific and cell-specific chromosomal copy numbers and markers [labeled in (A and B)] that could only be distinguished as such by the statistical analyses of the karyotype arrays shown in .
![Figure 6. Two individual karyotypes of the cen3tel clone PD166 at its origin (A) and after 848 generations of unselected growth in culture at PD1014 (B). About 64% of the chromosomes of the two karyotypes share PD166-specific copy numbers or are common markers (Table 4 and Fig. 5). The two karyotypes differ in generation-specific and cell-specific chromosomal copy numbers and markers [labeled in (A and B)] that could only be distinguished as such by the statistical analyses of the karyotype arrays shown in Figure 5.](/cms/asset/1f8b9d97-1ef3-4e16-b18c-381aed8445d9/kccy_a_10923720_f0007.gif)
Figure 7. (A) Karyotype of the immortal tumorigenic clone cen3telS2 PD84, which arose from an independent preneoplastic aliquot of the cen3tel mass culture 84 generations after it was transfected with overexpressed telomerase. (See above Results, “How does overexpressed telomerase induce…”) (B) Karyotype of cen3telS2 PD143, which supplanted the PD84 clone 59 generations after it was identified at PD84.
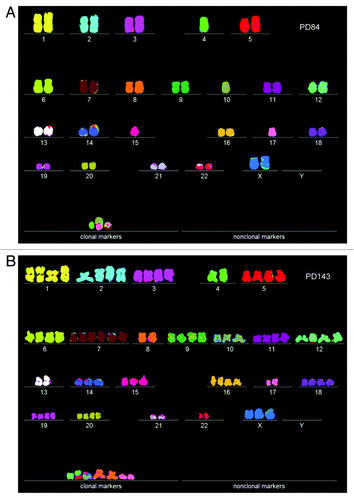
Table 5. Chromosome numbers and copy numbers of immortal, tumorigenic clones cen3telS2 PD84 and subclone cen3telS2 PD143
Figure 8. Karyotype arrays of the immortal, tumorigenic clones cen3telS2 PD84 (A) and a sub-clone cen3telS2 PD143 (B) based on data of . The cen3telS2 PD84 clone arose spontaneously in an independently growing preneoplastic aliquot of the cen3tel mass culture (from which also cen3tel PD166 originated) 84 generations after transfection with overexpressed telomerase. Upon further cultivation for 59 generations, the cen3telS2 PD84 clone was supplanted by a more transformed and faster growing variant, cen3telS2 PD143. Comparative karyotype arrays revealed that PD143 is a near-tetraploid, clonal variant of cene3teS2 PD84 ().
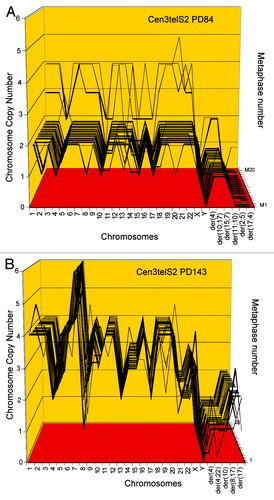
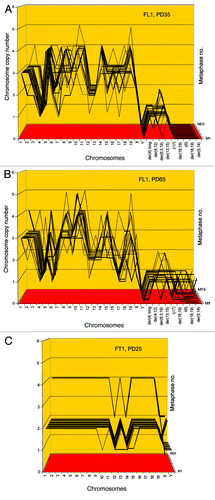
Figure 9. Karyotypes of immortal clones of SV40 virus-transformed lung cells, FL1 (A) and tail cells, FT1 (B) from a telomerase-deficient mouse (see also ). The transformed clones arose together with their new individual karyotypes 2–3 mo after infection of the cells with SV40 virus.
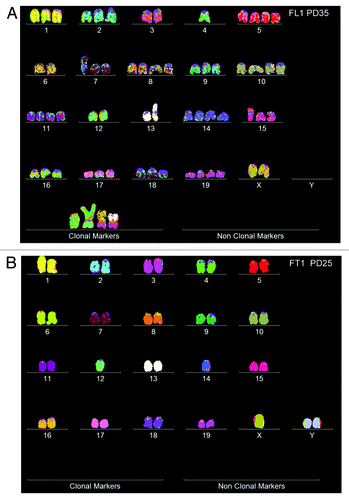
Figure 10. Karyotype arrays of immortal, neoplastic clones of SV40 virus-transformed lung cells, FL1 (A and B) and tail cells, FT1 (C) from a telomerase-deficient mouse. The clones appeared 3 mo after infection by SV40 tumor virus together with their new clonal and flexible karyotypes. Karyotype arrays of the lung-derived focal culture, FL1 were determined 35 and 65 generations after isolation of the focal culture. The arrays show that the FL1 karyotype was near-triploid with near clonal FL1-specific chromosome numbers and chromosome copy numbers (see also ). The karyotype array of the tail-derived focal culture, FT1 was determined 25 generations after isolation of the focal FT1 clone. The array shows that the FT1 karyotype was hypo-diploid with 94–100% clonal chromosome numbers and chromosome copy numbers.