Figures & data
Figure 1. Effects of transduction of various Ras mutants on Ras content, ROS level as determined by DCF fluorescence and sestrins genes expression in HF and HaCaT cell cultures. (A) Western blot analysis of Ras content; actin protein was analyzed as loading control. (B) Upper panel: average indices of total DCF fluorescence of 104 cells (in each case summarized data of three independent experiments are shown). HF (left) and HaCaT (right) cell sublines and cell cultures were analyzed 11–15 d after introduction of corresponding retroviral pLXSN-neo vectors. At the bottom, RT-PCR of SESN1, SESN2 and SESN3 mRNAs in HaCaT and HF expressing various Ras mutants for 11–15 d; tubulin mRNA was analyzed as loading control.
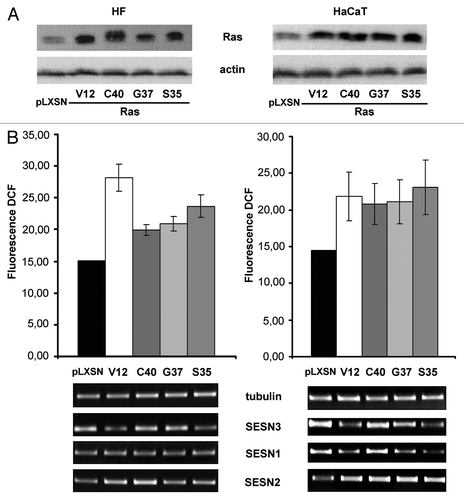
Figure 2. Reversion of Ras-induced changes in intracellular ROS content and sestrins genes expression in HF (left) and HaCaT (right) cell cultures. (A) Changes of ROS level on days 11–24 d after introduction of corresponding retroviral pLXSN-neo vectors. The average data of two independent experiments in each case are presented. (B) Changes in SESN3 mRNA level on days 11–24 after introduction of corresponding retroviral pLXSN-neo vectors (11–15 d/16–24 d mean that the average data shown on the figure were estimated by measuring SESN3 mRNA level during the indicated period of time). Relative quantities of SESN3 mRNA as estimated by densitometry of the results of RT-PCR experiments. The relative intensity of SESN3 bands was estimated as a ratio of SESN3 PCR-products to tubulin bands. The average results of two–three experiments are presented, means ± SE are given.
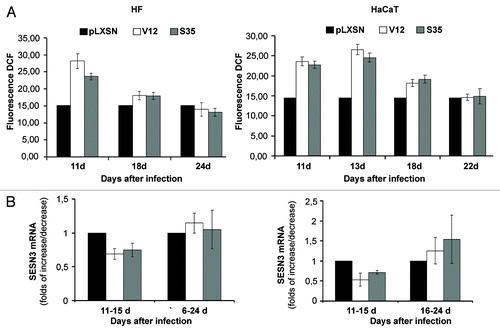
Figure 3. Effects of transduction of Ras mutants on HSF1 reporter activity (A) and HSF1 protein localization (B and C) in HF and HaCaT cell cultures. (A) The reporter lentiviral construct expressing luciferase gene under control of heat shock elements (HSE) was introduced into HF and HaCaT cells. Luciferase activity was measured 11–14 d after introduction of Ras mutants. (B) Nuclear, cytoplasmic fractions and total protein lysates were collected 11–14 d after Ras mutants introduction and were subjected to western blot analysis using anti-HSF1, anti-histone 3 (a nuclear marker), anti-actin (a cytoplasmic marker) antibodies. Actin protein was used as loading control for total protein lysates. (C) HSF1 immunofluorescence staining of HaCaT and HF cell cultures expressing RasV12 mutant or empty vector.
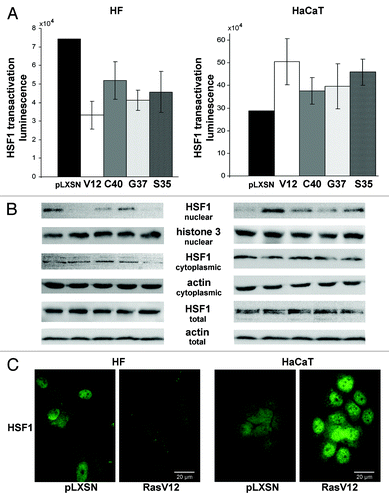
Figure 4. Effects of constitutively active (Act) and dominant-negative (dN) HSF1 mutants and shHSF1 on HSF1 reporter activity (A), ROS content (B) and sestrins mRNAs levels (C and D) in HF and HaCaT cells. In each case luciferase activity and DCF fluorescence were measured in three–five experiments; average data are presented. (C) RT-PCR of SESN1, SESN2 and SESN3 mRNAs; tubulin mRNA was analyzed as loading control. (D) The relative intensity of SESN3 bands was estimated as a ratio of SESN3 PCR-products to tubulin bands by densitometry of the results of four experiments, means ± SE are given.
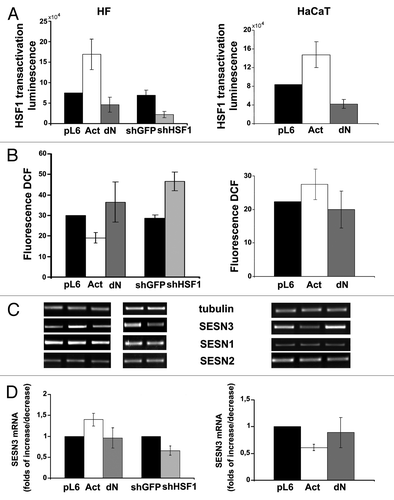
Figure 5. Effects of constitutively active (Act) and dominant-negative (dN) HSF1 mutants and shHSF1 on ROS content (A) and sestrins mRNAs levels (B and C) in HCT116 and HT1080 cells. In each case DCF fluorescence were measured in three-five experiments; average data are presented. (B) RT-PCR of SESN1, SESN2 and SESN3 mRNAs; tubulin mRNA was analyzed as loading control. (C) The relative intensity of SESN3 bands estimated according the results of four experiments, means ± SE are given.
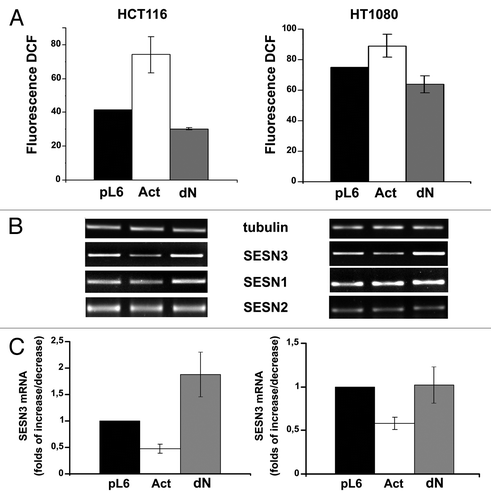
Figure 6. Ras introduction in HaCaT expressing shHSF1 doesn’t influence either ROS level or sestrins expression. (A) Cells expressing shHSF1 (or shGFP), RasV12 and shHSF1 together with RasV12 were subjected to western blot analysis using anti-HSF1, anti-panRas and anti-actin (as loading control) antibodies. (B) Average indices of total DCF fluorescence of 104 cells (in each case summarized data of three independent experiments are shown). (C) RT-PCR of SESN1, SESN2 and SESN3 mRNAs; tubulin mRNA was analyzed as loading control. (D) The relative intensity of SESN3 bands estimated according the results of four experiments, means ± SE are given.
Figure 7. Effects of different modifications of HSF1 function affecting SESN3 gene expression and ROS level on growth of HF, HaCaT and HCT116 cell cultures. (A) Effect of introduction of HSF1 constructs diminishing SESN3 expression and increasing ROS content. (B) Effect of introduction of HSF1 constructs increasing SESN3 expression and decreasing ROS level. In all cases, 2 x 104 cells were seeded into 6-well plates, and cell counts were performed until cell cultures reach monolayer using the hemocytometer (two wells per point). Typical result of one of three experiments is given; means ± SE are shown. At the bottoms average indices of total DCF fluorescence of 104 cells (in each case summarized data of three independent experiments are shown) (white) and of semi-quantitative RT-PCR of SESN3 mRNA (black).
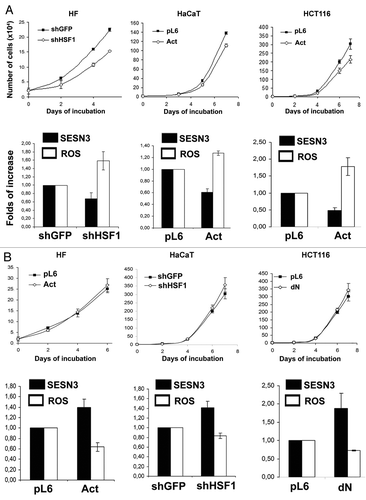
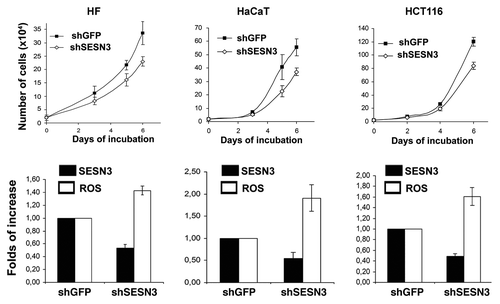
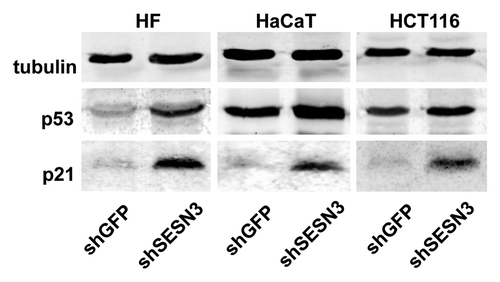
Figure 8. Effect of SESN3 downregulation caused by RNA interference on ROS level and growth of HF, HaCaT and HCT116 cell cultures. In all cases, 2 x 104 cells were seeded into 6-well plates, and cell counts were performed until cell cultures reach monolayer using the hemocytometer (two wells per point). Typical result of one of three experiments is given; means ± SE are shown. At the bottom, average indices of total DCF fluorescence of 104 cells (in each case summarized data of three independent experiments are shown) (white) and of semi-quantitative RT-PCR of SESN3 mRNA (black).