Figures & data
Figure 1. Creation of a 96-well macrowell array. (A) Assembly of macrowell comet array. Agarose gel with microwells is sandwiched between a glass substrate and a bottomless 96-well plate and sealed with mechanical force. Approximately 300 arrayed microwells comprise the bottom of each macrowell. (B) Loading and chemical dosing of cell samples in macrowell. One sample is loaded into each macrowell and cells settle by gravity into the arrayed microwells. The bottomless plate is removed in order to aspirate excess cells and enclose the cells in agarose. The bottomless plate is replaced in order to treat each macrowell with a chemical condition.
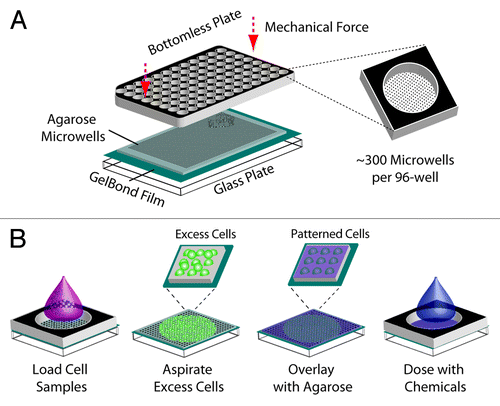
Figure 2. Arrayed microwell comet assay for detection of double-strand breaks. (A) Arrayed microwell comets from untreated TK6 human lymphoblasts and TK6 cells exposed to 100 Gy IR. Scale bar is 100 μm. (B) Comparison of irradiation dose response between traditional comet slides scored using commercial software and microwell comets scored using automated software. Each data point is the average of three independent experiments, where the median percent total DNA in tail (top), the median olive tail moment (middle), or the median tail length (μm) of 100 individual comets were used to represent the extent of DNA damage. Error bars represent the standard error of the mean from the four independent experiments. % DNA tail, R2 = 0.92 (CometChip) and R2 = 0.42 (traditional); olive tail moment, R2 = 0.97 (CometChip) and R2 = 0.63 (traditional); tail length (μm), R2 = 0.94 (CometChip) and R2 = 0.85 (traditional). (C) Bleomycin dose response conducted on macrowell platform with each data point representing the median tail length (μm) of at least 300 comets pooled from six macrowells.
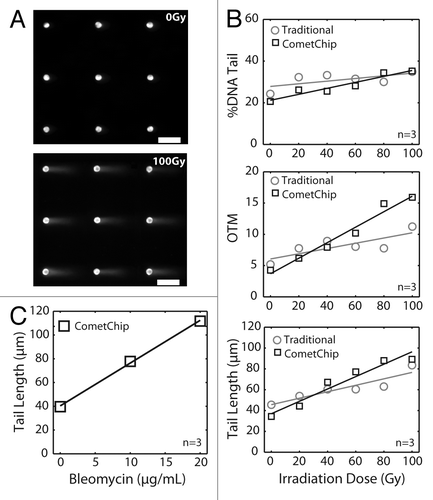
Figure 3. Statistical analysis and reproducibility of the CometChip compared with traditional comet slides. (A) IR dose response of TK6 cells loaded into different sized microwells. Each data point is the average of three independent experiments, where the median tail length of at least 100 individual comets was obtained in each experiment. Fluorescent images display 25, 30 and 45 μm diameter microwells filled with Syber Gold stained TK6 human lymphoblasts. Scale bar is 10 μm. (B) TK6 cells were irradiated with 100 Gy IR. Randomly selected 10, 50 or 100 individual comets on traditional glass slides or microwell comets on the CometChip were analyzed. Each data point is a single comet or microwell comet. Box plots show median tail length (μm) of the data set as the middle line and the lower and upper quartiles as the box. Whiskers show extent of furthest data points within 150% of interquartile range. (C) Slide-to-slide variability of traditional comet assay and macrowell-to-macrowell variability of 96-well format CometChip using TK6 cells exposed to 100 Gy IR. For traditional slides, medians of 30 traditional neutral comets from 10 slides are plotted. Mean comet length of 10 sides, 69 μm; coefficient of variation, ~20%. For CometChip, medians of at least 50 microwell comets from 10 macrowells of the CometChip are plotted. Mean comet length of 10 macrowells, 109 μm; coefficient of variation, ~5%.
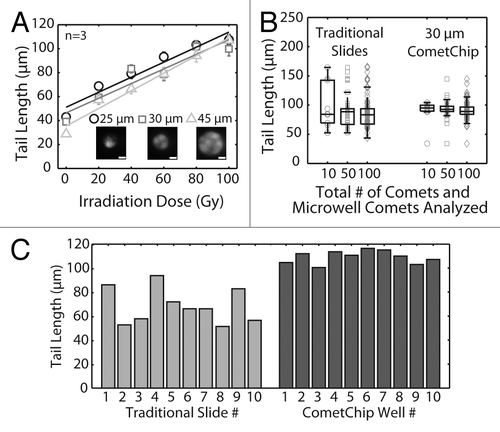
Figure 4. Evaluation of DNA repair kinetics of CHO-K1 (wild type), xrs6 (Ku80−/−) and irs20 (DNA-PKcs−/−) exposed to 100 Gy IR. All cell types and repair times conducted on a single CometChip with data representing median comet tail lengths (μm) from at least 50 comets. Error bars represent standard deviations of three independent experiments. Symbols indicate a significant difference compared with wild type according to t-test: *p < 0.05, **p < 0.005
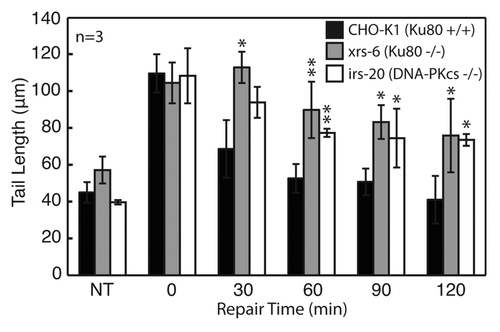
Figure 5. Comparison of neutral CometChip to γ-H2AX assay for repair kinetics. Wild type cells were exposed to 100 Gy IR. (A) All cell conditions and repair time points were conducted in triplicate macrowells of a single CometChip. Comet tail length (μm) values were normalized to peak wild type damage. (B) Repair kinetics were measured in triplicate over eight hours using a western blot version of the γ-H2AX assay. Error bars represent standard deviation of three replicate experiments.
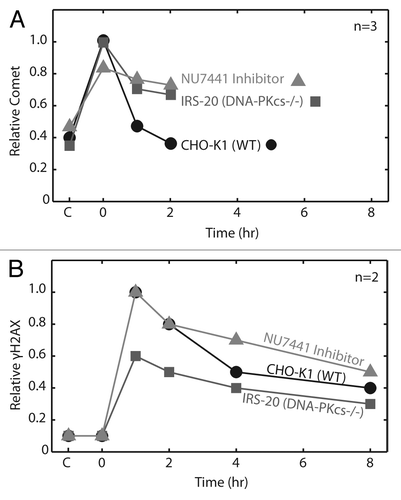
Table 1. DNA-PK inhibitors
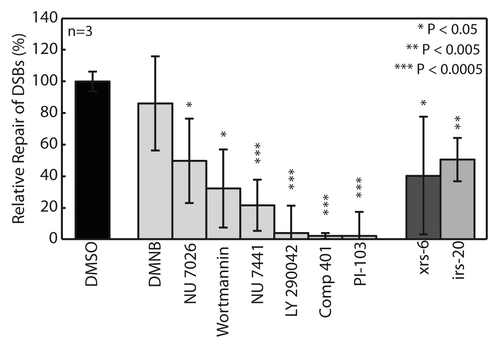
Figure 6. Evaluation of relative repair from 100 Gy IR after 1 h exposure to DNA-PK inhibitor library. CHO-K1 (wild type) cells pre-incubated for 1hr in 50 μM of each inhibitor. All conditions and controls, xrs6 (Ku80−/−) and irs20 (DNA-PKcs−/−), assayed in triplicate macrowells. Data and error bars represent averages and standard deviations of three independent experiments. Symbols indicate significance compared with wild type (DMSO) according to t-test: *p < 0.05, **p < 0.005, ***p < 0.0005.