Figures & data
Figure 1. Metabolic reprogramming from oxidative metabolism in fibroblasts to glycolysis in iPS cells. Representative serial nuclear magnetic resonance-based metabolomic footprinting of cell culture media indicates elevated glucose utilization and accumulation of lactate in iPS cells (A). High-throughput simultaneous measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) (B) indicates reduced oxygen utilization and greater reliance on glycolysis in iPS cells (C). Extracellular flux analysis in response to oligomycin, FCCP, and rotenone demonstrates an iPS cell specific mitochondrial profile characterized by reduced maximal respiration (D), reserve capacity (E) and ATP turnover (F). Values represent mean ± standard error, n = 8 per group. Significance was determined by Student t test. *P < 0.05 vs. MEF.
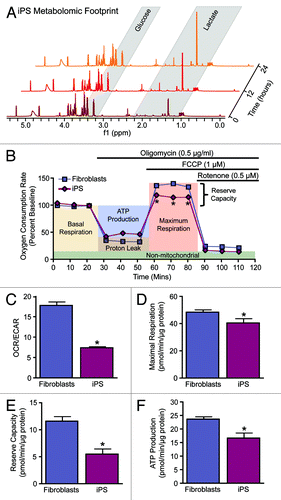
Figure 2. Nuclear reprogramming induced restructuring of the mitochondrial infrastructure. The mitochondrial anatomy and prevalence is remodeled during nuclear reprogramming as resolved by electron microscopy (arrows demarcate mitochondria) (A). iPS cells demonstrate mitochondrial membrane hyperpolarization compared with parental fibroblasts based upon mitochondrial accumulation of JC-1 (B).
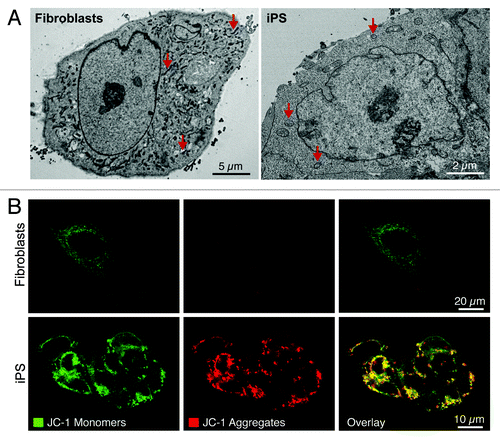
Figure 3. Nuclear reprogramming induced remodeling of the bioenergetic infrastructure. iPS cell-specific proteome is revealed in representative silver-stained, broad pH range (3–10) large format two-dimensional gels (A). Two-dimensional gel electrophoresis consistently resolved on the order of 550 protein species in fibroblast and iPS cell extracts, with densitometric quantification identifying a subset of 97 significantly altered protein spots (21% of the total resolved) following nuclear reprogramming, with significance determined by Student t test (P < 0.05) (B).
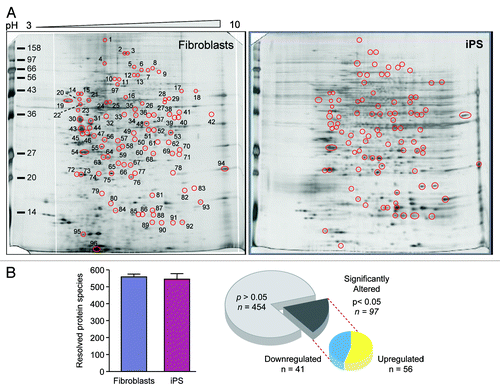
Figure 4. Pathway analysis interaction mapping clustered the upregulated subproteome into a fully connected network of 457 nodes and 3069 edges. Network degree distribution exhibits scale-free topology as defined by the relationship between node degree (k) vs. node degree distribution (P[k]), the proportion of all nodes at each specified degree (see Fig. S2A).Citation68,Citation69
![Figure 4. Pathway analysis interaction mapping clustered the upregulated subproteome into a fully connected network of 457 nodes and 3069 edges. Network degree distribution exhibits scale-free topology as defined by the relationship between node degree (k) vs. node degree distribution (P[k]), the proportion of all nodes at each specified degree (see Fig. S2A).Citation68,Citation69](/cms/asset/c7a4240a-4f10-470c-809d-606cbb53b7a7/kccy_a_10925509_f0004.gif)
Figure 5. Pathway analysis interaction mapping clustered the downregulated subproteome into a fully connected network of 392 nodes and 3237 edges. Network degree distribution exhibits scale-free topology as defined by the relationship between node degree (k) vs. node degree distribution (P[k]), the proportion of all nodes at each specified degree (see Fig. S2B).Citation68,Citation69
![Figure 5. Pathway analysis interaction mapping clustered the downregulated subproteome into a fully connected network of 392 nodes and 3237 edges. Network degree distribution exhibits scale-free topology as defined by the relationship between node degree (k) vs. node degree distribution (P[k]), the proportion of all nodes at each specified degree (see Fig. S2B).Citation68,Citation69](/cms/asset/7cf6815c-520b-4246-8e21-446631668ab3/kccy_a_10925509_f0005.gif)
Figure 6. Pathway analysis interaction mapping prioritizes canonical metabolic processes within the upregulated and downregulated iPS subproteomes. Ingenuity Pathway Analysis identified a number of overrepresented (P < 0.001) metabolic canonical pathways within the upregulated (A) and downregulated proteins arising from somatic cell reprogramming (B).
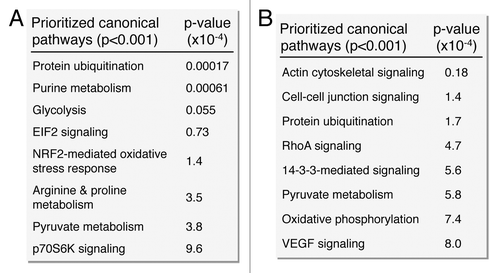
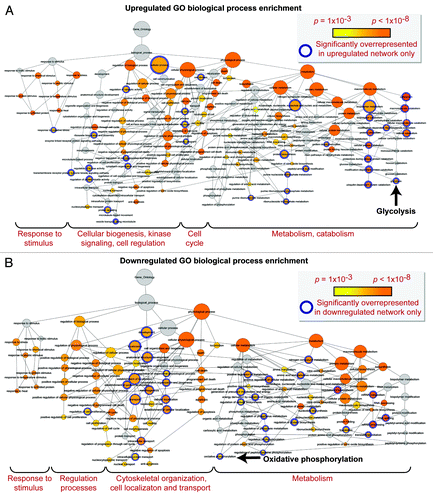
Figure 7. Gene ontology biological process prioritization within the upregulated and downregulated iPS cell subproteome networks. Unique to the upregulated network were 36 significantly overrepresented (P < 0.001) GO processes, including catabolic/metabolic functions mainly related to purine metabolism and glycolysis, cell cycle functions, and processes related to cell organization/biogenesis, kinase signaling, and an assortment of cell regulation functions (A). Unique to the downregulated network were 34 significantly overrepresented processes, associated with oxidative phosphorylation and several phosphate metabolism regulation and RNA related metabolic functions, as well as cytoskeletal structure/morphology and organization processes, cell localization, and transport (B).