Figures & data
Figure 1. DNA repair pathways of single strand DNA damages. Every day, cells are faced with thousand of damages involving base damages or bulky adducts, which have to be repaired to preserve cell function. Base damages induced by depurination or deamination are repaired by base excision repair, bulky adducts (in particular pyrimidine dimers induced by UV radiation) by nucleotide excision repair and base mismatch occurring during DNA replication by mismatch repair. Repair pathways involve first sensing of damage, followed by cleavage of damaged DNA, and synthesis of the correct DNA sequence using complementary strand as template.
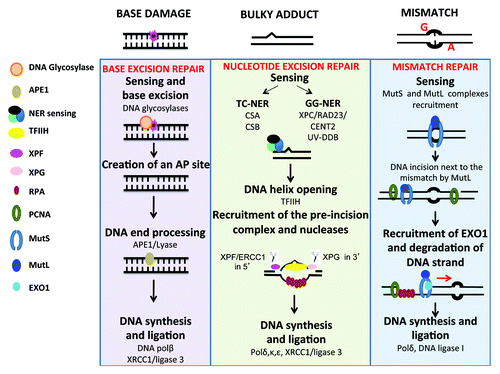
Figure 2. DNA repair pathways of double-strand DNA breaks. DNA double-strand breaks (DSB) occur in transcriptionally active or replicating cells in the presence of reactive oxygen species, ionizing radiation, or when the replication machinery encounters unrepaired single-strand damage. DSBs also enable DNA rearrangements in order to select for antigen-specific T or B lymphocytes. In addition, the main drugs used in cancer target cell replication machinery resulting in DSB and cell death. Two major pathways compete to repair DSBs. The accurate homologous recombination (HR) requires a duplicated sister chromatid and is active only at the end of S phase or in the G2 phase. Non-homologous end joining (NHEJ) is active in all phases of cell cycle and involved partial resection and ligation of DNA ends. The choice between HR and NHEJ in replicating cells is dictated by 53BP1 and BRCA1, which compete to bind DNA ends and block mutually. alt-NHEJ, alternative less accurate pathways, and SSA, single-strand annealing, can repair DSBs and may yield to DNA deletion, amplification, or translocation.
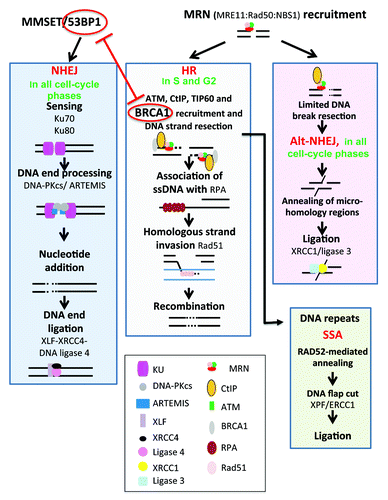
Figure 3. DNA repair of interstrand crosslinks (ICLs). Toxic compounds can create DNA Interstrand Crosslinks (ICLs), which impair transcription and replication machinery, and yield to cell death if unrepaired. ICL formation is the active principle of many cancer drugs, in particular alkylating agents. The removal of ICL involves the Fanconi anemia pathway, with sensing of ICL by FANCM, and then recruitment of protein complex resulting in ICL removal, creation of DSB, which is repaired by homologous recombination. In non-cycling cells, the initial sensing by FANCM is followed by ICL removal and DSB repair by poorly known mechanisms.
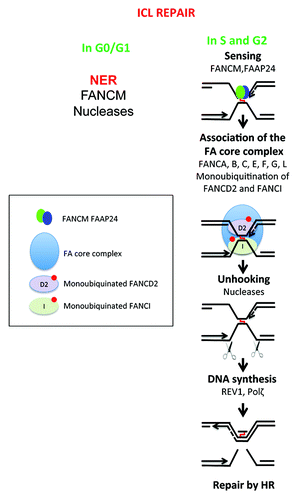
Figure 4. Double-strand DNA repair pathways in multiple myeloma. Genes coding for homologous recombination or non-homologous end joining pathways, whose promoter methylation (digit 1), polymorphism (digit 2), expression (digit 3), pathway activity (digit 4) are modified in multiple myeloma cells, are indicated. They are highlighted in red if the change is associated with a bad prognosis, in green with a good one, and blue in case of no prognostic value. The upwards or downwards arrows indicate an increased or decreased gene or protein expression or pathway activity. The existence of a truncated variant is indicated by digit 5.
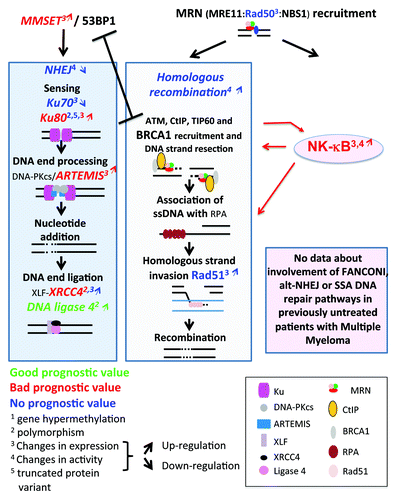
Figure 5. Single-strand DNA repair pathways in multiple myeloma. The changes in genes implicated in single-strand damage repair (BER, NER, or MMR) and modified in MM cells compare with healthy cells are highlighted. As indicated in , digit 1 indicates changes in promoter methylation, digit 2 the existence of polymorphisms, digit 3 changes in protein expression and digit 4 changes in activity. The upwards or downwards arrows indicate an increased or decreased gene expression or activity.
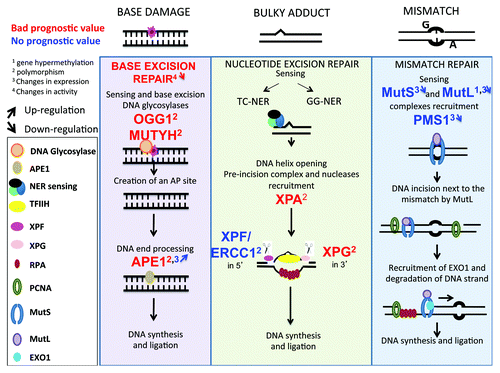
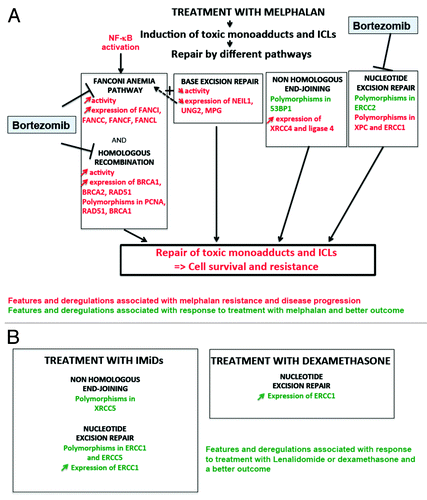
Figure 6. DNA repair pathways and drugs used to treat patients with multiple myeloma. (A) Velcade and Melphalan, 2 of the main drugs used to treat MM patients, modify DNA repair pathways in 2 opposite ways. Melphalan induces adducts and ICLs that need to be repaired mainly by the Fanconi anemia pathway and homologous recombination. Base excision repair (BER), non-homologous end joining and nucleotide excision repair (NER) pathways are also involved in ICLs repair. Changes in activity or expression of genes involved in these pathways or the existence of polymorphisms may activate and thus ease the repair of the lesions leading to cell survival and resistance to Melphalan. Some polymorphisms are associated with a better response after treatment. Changes in BER pathway can affect ICLs repair: an increased activity could help to repair the lesions but a decreased activity may also facilitate ICLs repair by the Fanconi anemia pathway by improving DNA accessibility. Velcade inhibits Fanconi anemia pathway, homologous recombination and NER. In red are summarized the features and deregulations associated with disease progression and resistance to melphalan, in green the features and deregulations associated with response to treatment and a better outcome after treatment with melphalan. (B) Very few data show a possible association between treatment with IMiDs and dexamethasone and DNA repair pathways. In green are summarized polymorphisms and changes in protein expression associated with a better outcome after treatment with dexamethasone or IMiDs (lenalidomide, thalidomide, and pomalidomide).