Figures & data
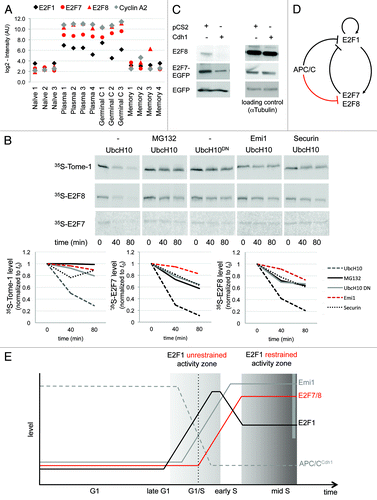
Figure 1. Using the B-cell lineage for identifying in vivo cell cycle regulators. (A) Splenic plasmablast and germinal center B cells are highly proliferative (red), whereas naïve and memory B-cells hardly, if ever, divide (blue). (B) Genes whose expression is switched on in both plasmablast and germinal center B cells, and switched off in both naïve and memory B cells, are likely to regulate the mammalian cell cycle and to promote proliferation in vivo. Genes with the reverse expression pattern are likely to promote antiproliferative processes in vivo. Overall, there are 16 combinatorial patterns of gene expression (illustrated as squares), of which 1 is the cell cycle pattern (red) and 1 is the antiproliferative pattern (blue).
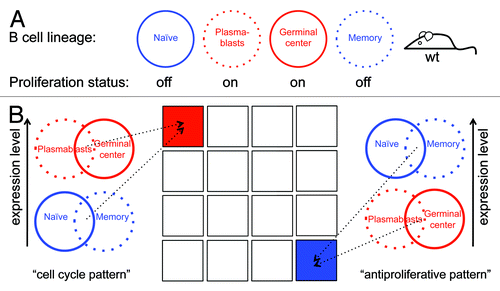
Figure 2. Defining the transcriptome signature of cell proliferation. (A) Normalized expression levels (log2-intensity, arbitrary units [AU]) of the cyclins B1 (CCNB1), B2 (CCNB2), and E2 (CCNE2) in the 4 B-cell types (see ), according to GEO data set GSE4142. (B) The complete GSE4142 data set was analyzed to identify genes most closely correlated with the cell cycle pattern (see colored bar). We complemented the analysis by GSEA with default settings, indicating significant enrichment (nominal P value < 0.001) of the cell cycle gene set, as defined by Whitfield et al.Citation1 (C) A heat map showing differentially expressed genes in proliferating (plasmablast and GC) vs. resting (naïve and memory) B cells. We used the ComparativeMarkerSelection module (GenePattern) to identify differentially expressed genes based on SNR with co-variance >0.4 and FDR Q value <0.02 in the comparison of proliferating vs. resting phenotypes. Eighty-three genes are depicted as cell cycle genes, of which 30 are known APC/C targets (marked with black triangles). (D) Top 10 GO term categories (ordered by P value; all P values <0.000 01) over-represented in our list of 83 cell cycle genes (AmiGO search tool; default parameters).
![Figure 2. Defining the transcriptome signature of cell proliferation. (A) Normalized expression levels (log2-intensity, arbitrary units [AU]) of the cyclins B1 (CCNB1), B2 (CCNB2), and E2 (CCNE2) in the 4 B-cell types (see Fig. 1A), according to GEO data set GSE4142. (B) The complete GSE4142 data set was analyzed to identify genes most closely correlated with the cell cycle pattern (see colored bar). We complemented the analysis by GSEA with default settings, indicating significant enrichment (nominal P value < 0.001) of the cell cycle gene set, as defined by Whitfield et al.Citation1 (C) A heat map showing differentially expressed genes in proliferating (plasmablast and GC) vs. resting (naïve and memory) B cells. We used the ComparativeMarkerSelection module (GenePattern) to identify differentially expressed genes based on SNR with co-variance >0.4 and FDR Q value <0.02 in the comparison of proliferating vs. resting phenotypes. Eighty-three genes are depicted as cell cycle genes, of which 30 are known APC/C targets (marked with black triangles). (D) Top 10 GO term categories (ordered by P value; all P values <0.000 01) over-represented in our list of 83 cell cycle genes (AmiGO search tool; default parameters).](/cms/asset/9d45e314-fb09-4759-b685-ed680240de03/kccy_a_10926030_f0002.gif)
Figure 3. Profiling gene expression in proliferating vs. resting B cells reveals in vivo regulators of cell differentiation and survival. (A) A heat map showing the expression of 28 differentially expressed genes in proliferating and resting B cells (for more details, see ; Fig. S1). Gene functions are depicted (see Table S2 for references). (B) The top 7 GO term categories (ordered by P value) over-represented in the genes listed in (A) (AmiGO search tool; default parameters; all P values are below 0.008 5).
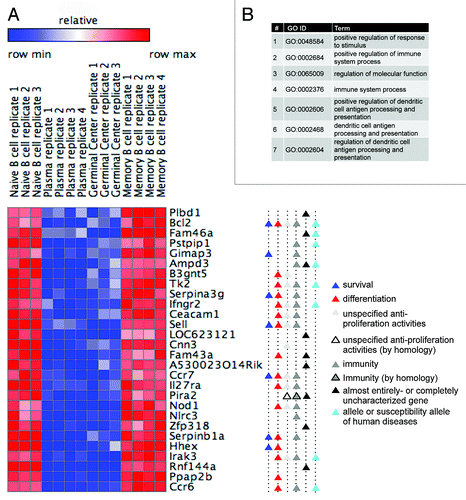
Figure 4. APC/CCdh1 targets E2F7 and E2F8 for degradation. (A) Normalized expression levels of E2F1, E2F7, E2F8, and cyclin A2 (CCNA2) in the depicted B-cell types, according to GEO database GSE4142. (B) The proteins XlTome-1, hE2F7, and hE2F8 were expressed in rabbit reticulocytes supplemented with 35S-methionine, and incubated in G1 phase S3 cell extracts supplemented with wt or dominant negative (DN) Ubch10 and either buffer, MG132, the C-terminus of Emi1, or unlabeled Securin. Time-dependent degradation was assayed by SDS-PAGE and autoradiography (top), and quantified (bottom). (C) HEK293 cells were co-transfected with Cdh1 or empty vector (pCS2) and with either E2F8, or E2F7-EGFP, or EGFP, at a 4:1 ratio. After 30 h, cells were harvested for western blotting with E2F8, GFP (experimental control), and tubulin (loading control) antibodies. (B and C) are representative experiments. (D and E) Model for the interplay between APC/CCdh1 and E2F1, E2F7, and E2F8. APC/CCdh1 regulates the proteolysis of both E2F1 and its repressors, E2F7 and E2F8 (B and C) during G1-phase. The model proposes that a delicate difference in the timing of proteolysis of the activating vs. repressive E2F transcription factors controls the G1/S transition of the mammalian cell cycle (see main text for further information/discussion).