Figures & data
Figure 1. PDGF reduces HDAC11 mRNA abundance in Balb/c-3T3 cells. (A) Density-arrested Balb/c-3T3 cells received 10 ng/ml PDGF and 10% PPP for 1, 3, or 7 h. A microarray analysis was performed as described in “Materials and Methods”. (B) Density-arrested Balb/c-3T3 cells were co-treated with 10 ng/ml PDGF and 10% PPP for the indicated times. Amounts of HDAC11 mRNA were determined by RT-qPCR. Data are expressed as percentage of control (no treatment, diagonal bar). Error bars show standard deviation. (C) Density-arrested Balb/c-3T3 cells received 10 ng/ml PDGF (left panel) or 10% PPP (right panel) for the indicated times. Amounts of HDAC11 mRNA were determined by RT-qPCR. Data are expressed as percentage of control (no treatment, diagonal bar). Error bars show standard deviation.
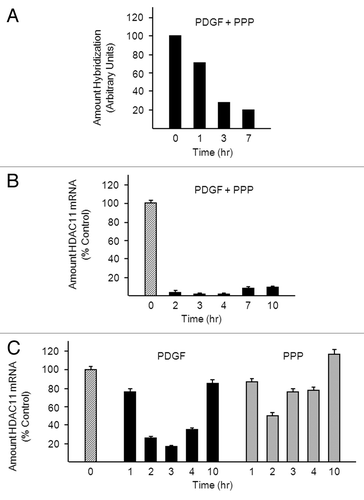
Figure 2. PDGF-mediated downregulation of HDAC11 mRNA requires protein synthesis. (A) Density-arrested Balb/c-3T3 cells received 10 ng/ml PDGF, 50 ng/ml EGF, 40 ng/ml IGF-1, or 5 μg/ml insulin for 1, 2, or 3 h. Amounts of HDAC11 mRNA were determined by RT-qPCR. Data are expressed as percentage of control (no treatment, diagonal bar). Error bars show standard deviation. (B) Density-arrested Balb/c-3T3 cells received 1 μM LY294002 or 10 μM U0126 for 25 min followed by 10 ng/ml PDGF for 2.5 h. Left panel: Amounts of HDAC11 mRNA were determined by RT-qPCR. Data are expressed as percentage of control (no treatment, diagonal bar). Error bars show standard deviation. Right panel: Amounts of phosphophorylated AKT (pAKT) and phosphorylated ERK (pERK) were determined by western blotting of cell extracts with phospho-specific antibodies. (C) Density-arrested Balb/c-3T3 cells received 10 ng/ml PDGF, 20 µg/ml cycloheximide (CHX), or both for 0.5, 1 or, 2 h. Amounts of HDAC11 mRNA were determined by RT-qPCR. Data are expressed as percentage of control (no treatment, diagonal bar).
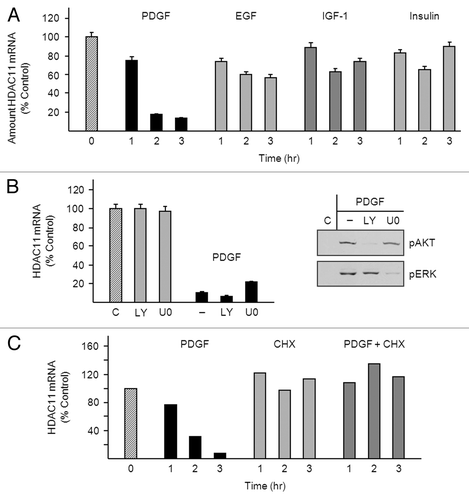
Figure 3. PDGF destabilizes the HDAC11 transcript in a transcription-dependent manner. (A) Density-arrested Balb/c-3T3 cells received 10 ng/ml PDGF, 50 μM DRB or both for the indicated times. Amounts of HDAC11 mRNA were determined by RT-qPCR. Decay curves were generated by linear regression. (B) Growing Balb/c-3T3 cells (approximately 80% confluent) were co-transfected with pRL-CMV and either pGL3-Basic or pGL3-HDAC11. Cells received 10 ng/ml PDGF 38 h after transfection (0 h) and were harvested at 3, 6, or 9 h thereafter for determination of luciferase activity. Cells were quiescent and confluent at the time of PDGF addition. Firefly luciferase activity (pGL3) is normalized to Renilla luciferase activity (pRL-CMV) (RLU1/RLU2). Error bars show standard deviation.
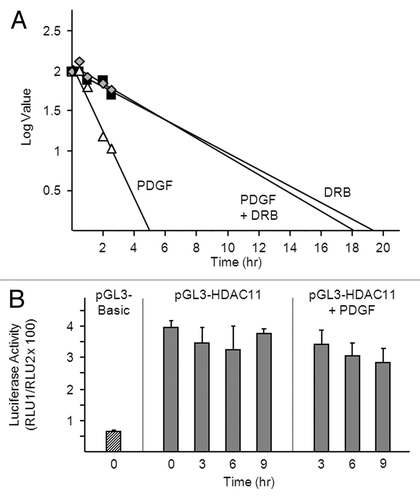
Figure 4. Accumulation of HDAC11 mRNA in confluent Balb/c-3T3 but not clone 2 cells (A) Amounts of HDAC11 mRNA were determined by RT-qPCR on the days indicated (top panel). Percentages of cells in G0/G1, S, and G2/M were determined by FACS analysis of propidium iodide-stained cells (bottom panel). Error bars show standard deviation. (B) Cultures were photographed on the days indicated. Magnification is 10×. (C) Sparse (day 2) and confluent (day 7) Balb/c-3T3 cells received 10 ng/ml PDGF for 3 h. Amounts of HDAC11 mRNA were determined. Error bars show standard deviation.
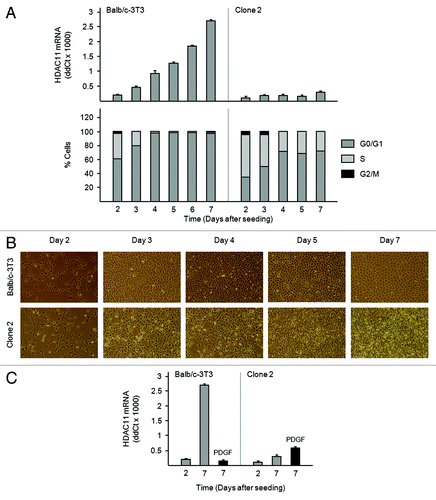
Figure 5. The HDAC11 promoter is active in Balb/c-3T3 but not clone 2 cells. (A) Proliferating Balb/c-3T3 and clone 2 cells (approximately 80% confluent) were transfected with pRL-CMV (for normalization) and either pGL3-Basic (–) or pGL3-HDAC11 (+). Cells were harvested 38 h later for determination of luciferase activity and were confluent at the time. Error bars show standard deviation. Data are expressed as fold increase in HDAC11 promoter activity. RLU1/RLU2 values for pGL3-Basic in Balb/c-3T3 and clone 2 cells were 0.011 and 0.035, respectively. (B) Genomic DNA was isolated from confluent Balb/c-3T3 and clone 2 cells, and CpG island methylation was determined as described in “Materials and Methods”.
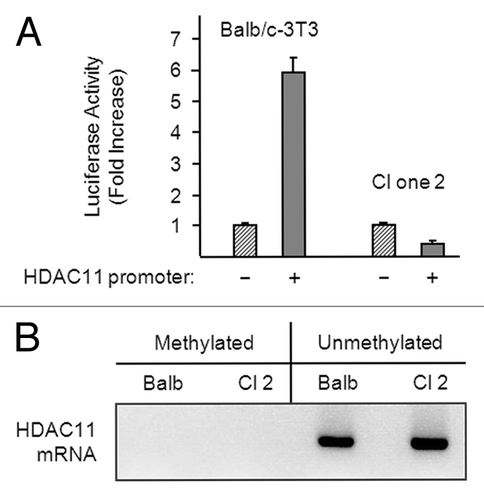
Figure 6. Serum deprivation increases amounts of HDAC11 mRNA in Balb/c-3T3 cells. (A–C) Sparse, cycling cells in 10% serum were refed with medium containing 10% or 0.5% serum (day 0). (A) Cultures were photographed on the days indicated. Magnification is 10×. (B) Percentages of cells in G0/G1, S, and G2/M were determined by FACS analysis of propidium iodide-stained cells. (C) Amounts of HDAC11 mRNA were determined by RT-qPCR on the days indicated. Error bars show standard deviation.
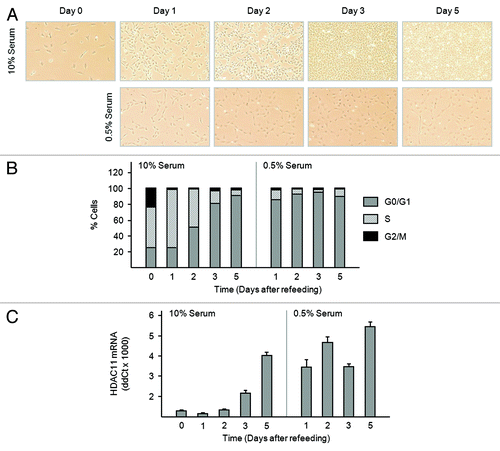
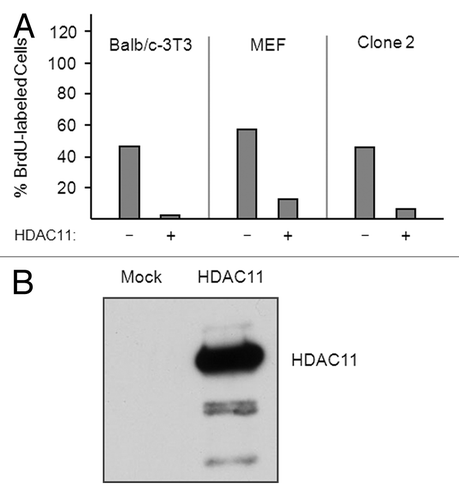
Figure 7. Overexpression of HDAC11 inhibits the cell cycle progression of nontransformed and transformed fibroblasts. (A) Sparse proliferating Balb/c-3T3, MEFs, and Clone-2 cells were transfected with plasmids encoding Flag-tagged HDAC11 and GFP. Thirty hours after transfection, cells were pulsed with 30 μM BrdU for 1 h and sorted for GFP expression. The percentage of BrdU-labeled cells was determined for cells expressing (+) or not expressing (–) ectopic proteins. (B) Balb/c-3T3 cells were mock transfected or transfected with a plasmid encoding HDAC11. Cell extracts were western blotted with a monoclonal antibody to HDAC11 prepared for us by Abpro Labs.